
The Picard Pipeline
Sequencing Pipeline Informatics at Broad

• What is Picard?
• Picard in context
• What Picard produces and how to access it
• Metrics, Metrics, Metrics

• A set of tools for processing and analyzing next generation
sequencing data
– Many of which are released publicly
• A set of pipelines that process all Illumina sequence data
generated at Broad

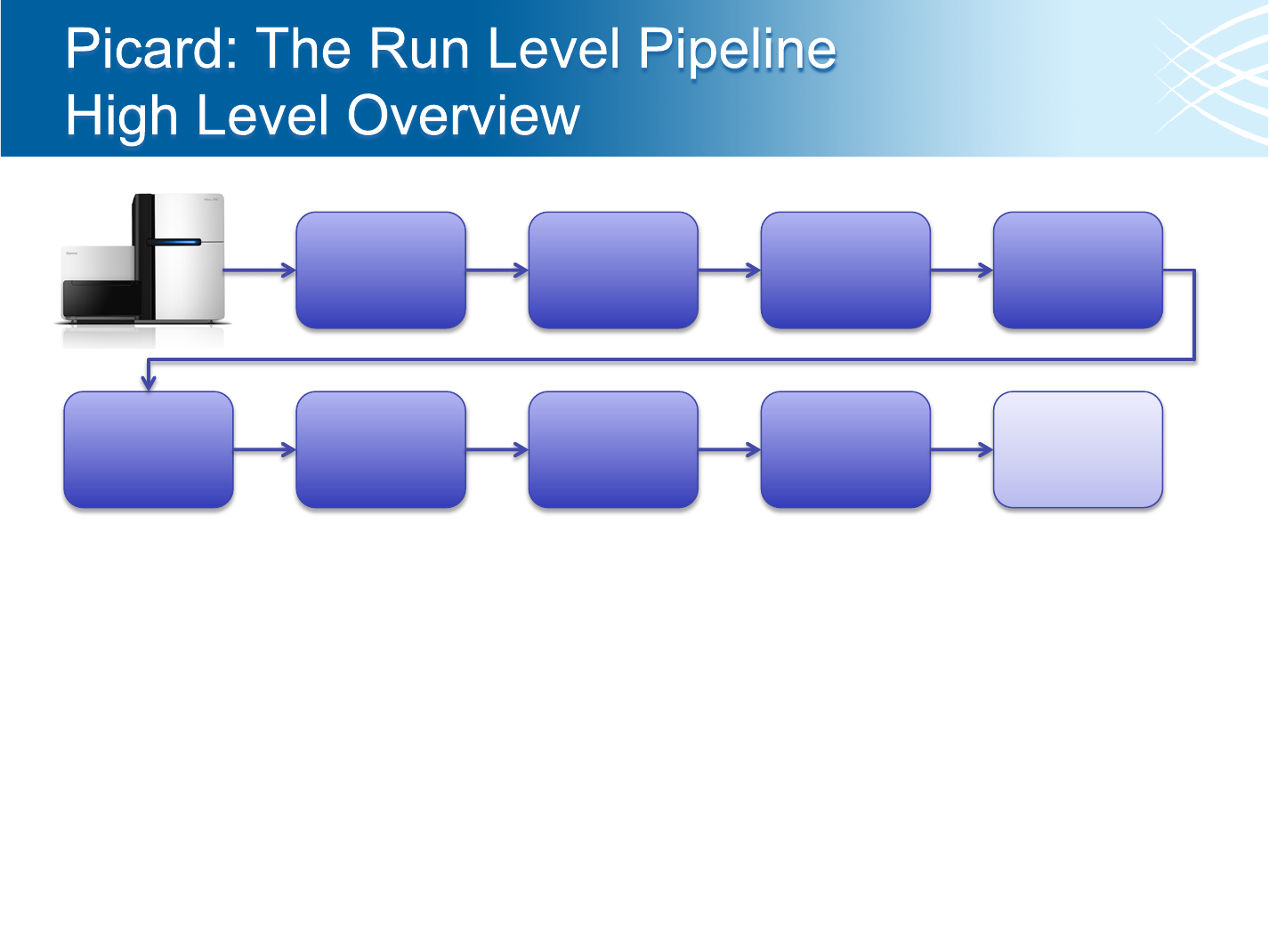
Extract
Illumina Data
to Standard
Format
Align reads
with BWA or
MAQ
Mark
Duplicate
Reads
Re-align
reads around
known indels
Calibrate
Quality
Scores
Collect
Metrics about
Libraries and
Run
Verify Sample
Identity
Run Triage
• Adapter trimming/marking happens during data extraction from Illumina (information is
used during alignment)
• Indexed runs are de-multiplexed during extraction and each index/sample processed
independently
• Recalibration only performed for references with dbSNP
Aggregation
Pipeline
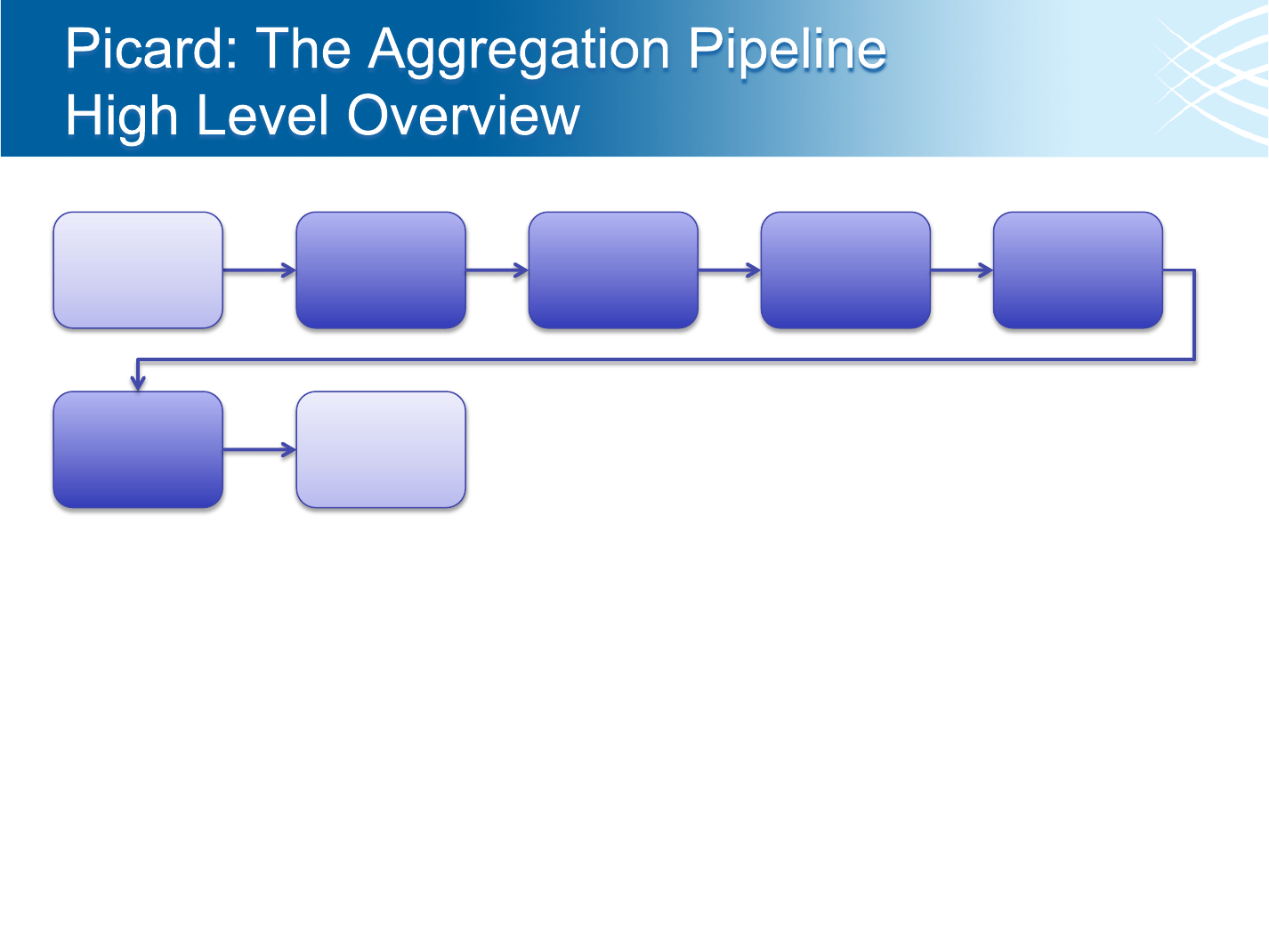
Merge all
data for each
library
Mark
Duplicate
Reads per
library
Collect
Metrics per
library
Merge all
libraries for a
sample
Collect
Metrics about
the Sample
• A single BAM file is created per Sample (within the context of a project)
• Aggregations are started after data is processed or re-processed through the run-level
pipeline (after a 12 hour “quiet period”)
• Outdated aggregations are kept for 2 weeks after newer aggregations are completed
Downstream
pipelines and
analysts
Run Level
Pipeline
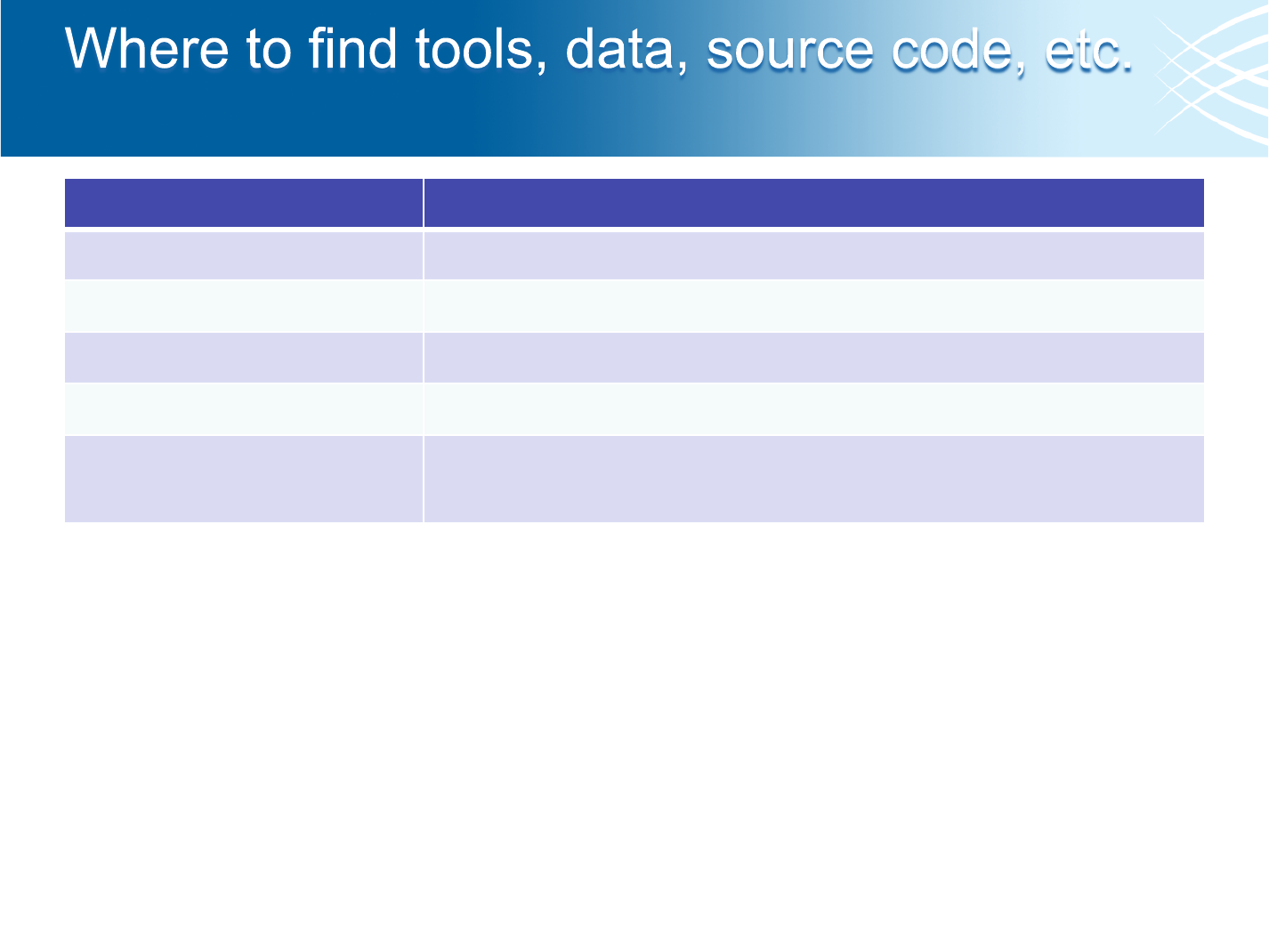
What
Where
Pipeline Outputs
/seq/picard/{flowcell}
Aggregation Outputs
/seq/picard_aggregation/{project}/{sample}
Picard Binaries
/seq/software/picard/current/bin
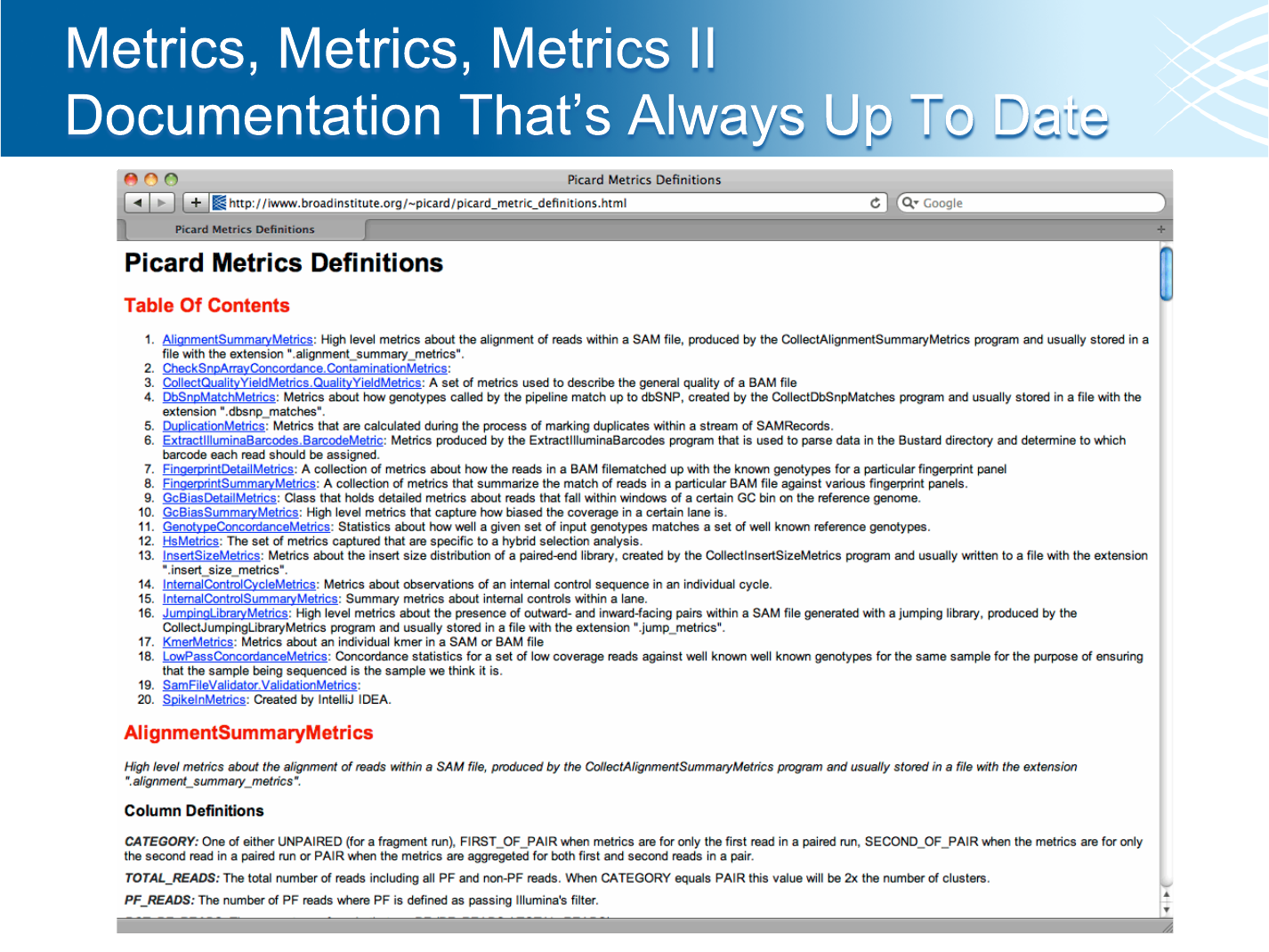
Metrics Documentation
http://iwww/~picard/picard_metric_definitions.html
Source Code
https://svn.broadinstitute.org/picard/trunk
https://picard.svn.sourceforge.net/svnroot/picard/trunk
• And coming soon – BASS
• Programmatic access to BAM files in BASS available
• Web page to access BAM files in BASS under construction

• All primary data is delivered in BAM format, which includes basecalls
(the reads), quality scores, alignment data, etc.
• BAM files processed through Picard always contain all reads,
including:
– All unaligned reads (marked as unmapped)
– All duplicate reads (marked as duplicates)
– All “non-PF” reads (marked as failing vendor quality)
• The pipeline generates tons of metrics!
• And we have tools to generate even more than run in the pipeline
• Please check before re-inventing the wheel
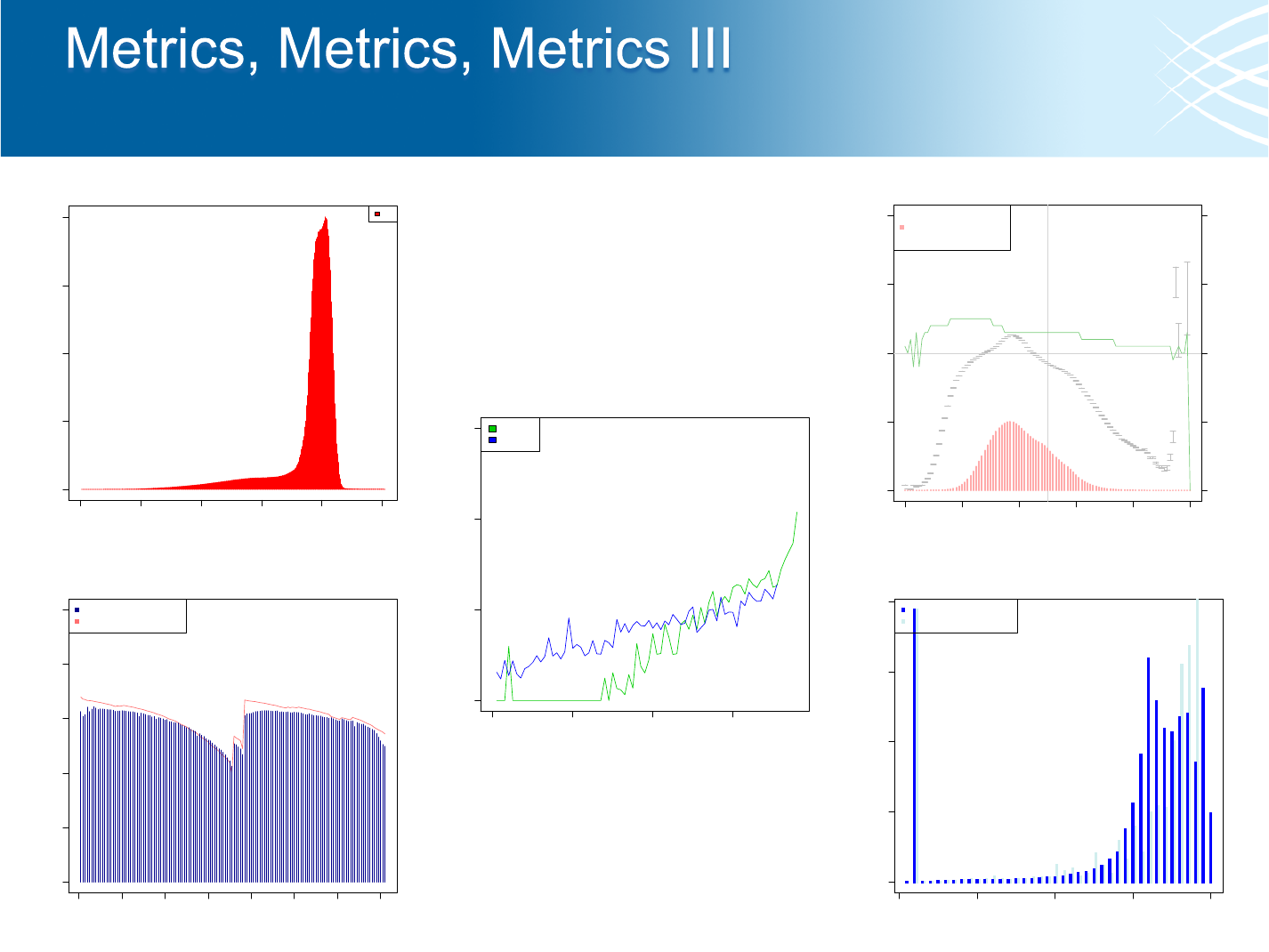
Internal Control Metrics
Quality Calibration Data
Alignment Summary Metrics
GC Bias Metrics
Quality By Cycle
Quality Distribution
Duplication Metrics
Insert Size Metrics
Low Pass Concordance
Hybrid Selection Metrics
SNP Fingerprint
Jumping Library Metrics
dbSNP Concordance
Quality/Yield Metrics
Barcode Metrics
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
!
0 20 40 60 80 100
0.0 0.5 1.0 1.5 2.0
20AL7ABXX.1 GC Bias Plot
Total clusters: 124,301,048, Aligned reads: 206,073,969
GC% of 100 base windows
Fraction of normalized coverage
0 10 20 30 40
Mean base quality
!
−
Normalized Coverage
Windows at GC%
Base Quality at GC%
0 100 200 300 400 500
0 500000 1000000 1500000 2000000
20AL7ABXX.1.aligned.duplicates_marked.bam Insert Size Histogram
Insert Size
Count
FR
0 20 40 60 80 100 120 140
0 10 20 30 40 50
209KJABXX.1.aligned.duplicates_marked.bam Quality By Cycle
Cycle
Mean Quality
Mean Quality
Mean Original Quality
0 10 20 30 40
0.0e+00 5.0e+08 1.0e+09 1.5e+09 2.0e+09
209KJABXX.1.aligned.duplicates_marked.bam Quality Score Distribution
Quality Score
Observations
Quality Scores
Original Quality Scores
0 20 40 60
209KJABXX.1.unmapped.bam Total (n=481735) IC Error Rate by Cycle
Cycle
Error Rate (log axis)
0.001 0.010 0.100 1.000
Read 1
Read 2

• Integrate GATK Unified Genotyper in single-sample mode
• Customized pipeline for cDNA/RNA sequencing
• Yet more sample identity/validity checking
