Vol.:(0123456789)
1 3
Histochemistry and Cell Biology
https://doi.org/10.1007/s00418-023-02209-1
ORIGINAL PAPER
OME‑Zarr: acloud‑optimized bioimaging file format withinternational
community support
JoshMoore
1
· DanielaBasurto‑Lozada
2
· SébastienBesson
3
· JohnBogovic
4
· JordãoBragantini
5
·
EvaM.Brown
6
· Jean‑MarieBurel
3
· XavierCasasMoreno
7
· GustavodeMedeiros
8
· ErinE.Diel
9
·
DavidGault
3
· SatrajitS.Ghosh
10
· IlanGold
11
· YaroslavO.Halchenko
12
· MatthewHartley
13
·
DaveHorsfall
2
· MarkS.Keller
11
· MarkKittisopikul
4
· GaborKovacs
14
· AybükeKüpcüYoldaş
13
·
KojiKyoda
15
· AlbaneleTournoulxdelaVillegeorges
16
· TongLi
17
· PriscaLiberali
8
· DominikLindner
3
·
MelissaLinkert
9
· JoelLüthi
8
· JeremyMaitin‑Shepard
18
· TrevorManz
11
· LucaMarconato
19
·
MatthewMcCormick
20
· MerlinLange
5
· KhaledMohamed
3
· WilliamMoore
3
· NilsNorlin
21
·
WeiOuyang
7
· BugraÖzdemir
22
· GiovanniPalla
23
· ConstantinPape
24
· LucasPelkmans
25
·
TobiasPietzsch
4
· StephanPreibisch
4
· MartinPrete
17
· NormanRzepka
16
· SameeulSamee
26
·
NicholasSchaub
27
· HythemSidky
26
· AhmetCanSolak
5
· DavidR.Stirling
9
· JonathanStriebel
16
·
ChristianTischer
28
· DanielToloudis
6
· IsaacVirshup
23
· PetrWalczysko
3
· AlanM.Watson
29
·
ErinWeisbart
30
· FrancesWong
3
· KevinA.Yamauchi
31
· OmerBayraktar
17
· BethA.Cimini
30
·
NilsGehlenborg
11
· MuzlifahHania
17
· NathanHotaling
27
· ShuichiOnami
15
· LoicA.Royer
5
·
StephanSaalfeld
4
· OliverStegle
19
· FabianJ.Theis
23
· JasonR.Swedlow
3
Accepted: 16 May 2023
© The Author(s) 2023
Abstract
A growing community is constructing a next-generation file format (NGFF) for bioimaging to overcome problems of scal-
ability and heterogeneity. Organized by the Open Microscopy Environment (OME), individuals and institutes across diverse
modalities facing these problems have designed a format specification process (OME-NGFF) to address these needs. This
paper brings together a wide range of those community members to describe the cloud-optimized format itself—OME-
Zarr—along with tools and data resources available today to increase FAIR access and remove barriers in the scientific
process. The current momentum offers an opportunity to unify a key component of the bioimaging domain—the file format
that underlies so many personal, institutional, and global data management and analysis tasks.
Keywords FAIR· Community· Bioimaging· Data· Cloud· Format
Introduction
The exchange of scientific data is one of the key hallmarks
of scientific practice in the twenty-first century. In 2016
Wilkinson and colleagues provided guidelines for making
scientific data findable, accessible, interoperable, and reus-
able (FAIR) that provide a foundation for future scientific
discoveries through data integration, reanalysis and the
development of new analytic tools (Wilkinson etal. 2016).
In the case of biological and biomedical imaging (collec-
tively, “bioimaging”), the size, complexity and heterogeneity
of datasets present several impediments towards that goal,
the most immediate of which are the specification and con-
struction of data formats that can meet the requirements of
FAIR data (Könnecke etal. 2015).
Any format must support both the pixel measurements that
are the core of bioimaging data as well as relevant imaging
metadata. Specifications that enable storage of experimental,
acquisition, and analytic metadata are necessary. The imple-
mentation of metadata specifications must be both flexible
and scalable to handle the large and heterogeneous volumes
of analytic metadata generated, for example the definition of
the segmentations and annotations on individual cells and tis-
sues that are quite common in biological imaging workflows.
Critically, the set of formats available to end users must sup-
port local data storage (laptops, desktop computers, etc.) as
Extended author information available on the last page of the article

Histochemistry and Cell Biology
1 3
well as cloud-based storage that is becoming more heavily
used as dataset volumes grow.
Previously, the Open Microscopy Environment (OME)
developed OME-TIFF as an open-source file format in
bioimaging. Accompanied by reference software imple-
mentations, OME-TIFF is primarily for use in fluorescence
imaging workflows and has recently been updated to enable
whole slide imaging technologies (Besson etal. 2019).
This format combines the fundamentally 2D TIFF format
with metadata cast in XML in the TIFF header. Its struc-
ture makes it appropriate for many applications, where the
plane-based access pattern is appropriate.
For bioimaging applications that require large non-pla-
nar access to volume data, e.g., arbitrary slicing from user-
defined angles, a more sophisticated “chunking” of the data
is required that defines how data is stored in accessible and
reasonable subsections. This means that large, multi-Giga-
byte up to Petabyte bioimaging datasets are not accessed all
at once but can be accessed in reasonably sized planes or
sub-volumes. In the case of TIFF, the chunk is a tile of the
2D plane allowing data access across time-lapse series or
3D volume. N-dimensional formats like HDF5 (“Hierarchi-
cal Data Format”) provide much more flexibility and allow
chunking across different dimensions chosen by the user.
While TIFF and HDF5 are well established, the chunking
strategies depend on fast random access to the entire file that
is common in laptops, desktop machines and large cluster
file systems, but is not provided by large scalable cloud-
based object storage.
Over the last few years, a new data format, Zarr
1
, has
been developed for the storage of large N-dimensional
typed arrays in the cloud. The Zarr format is now heavily
adopted across many scientific communities from genomics
to astrophysics (Miles etal. 2023). Zarr stores associated
metadata in JSON and binary data in individually reference-
able “chunk”-files, providing a flexible, scalable method for
storing multidimensional data. In 2021, OME published the
first specification and example uses of a “next-generation file
format” (NGFF) in bioimaging using the Zarr format (Moore
etal. 2021). The first versions of this format, OME-Zarr,
focused on developing functionality that tests and demon-
strates the utility of the format in bioimaging domains that
routinely generate large, metadata-rich datasets—high con-
tent screening, digital pathology, electron microscopy, and
light sheet imaging.
The discussions necessary to arrive at these specifications
have also presented an opportunity to build a coherent devel-
opment community under the OME-NGFF umbrella, combin-
ing a growing range of use cases and requirements with an
open, transparent, but technically valid development process.
The result has been a thriving community based on open
development and open-source principles (Rueden etal. 2019).
This open, collaborative approach has been essential to tackle
the addition of complex additional metadata to OME-Zarr.
This was important as neither TIFF nor HDF5 has specifica-
tions for many of the derived data types that are generated in
an analysis workflow, e.g., regions of interests (ROIs), labels
and other derived data which are crucial in modern analy-
sis workflows. In most cases accessory files are generated to
handle these limitations but as data volumes grow, these cre-
ate additional problems for management and linkage of data.
Using the established development process, this functionality
was first formally adopted into the OME-NGFF specification,
then added to the OME-Zarr implementation, but can equally
be applied to other formats like HDF5 in the future.
In this paper, we review the current status of the OME-
Zarr format and focus on resources that are now available
to users for creating, accessing, and visualizing data stored
in OME-Zarr (Fig.1). This report is timely as we have seen
a rapid expansion in tools that support OME-Zarr since the
first publication. We also report on the growth of adoption
of OME-Zarr in public data repositories. This survey by
active members of the OME-NGFF community is meant to
provide an update on the status of the ecosystem that has
grown around the format and the development community
that is developing and releasing tools that can be used by the
broader bioimaging community.
Growth ofacompatible solution
The development of a common format is not a light under-
taking. Historical approaches to address challenges of scale
most often offer a problem-specific and highly-optimized
solution, and do not generalize to the wider bioimaging
community, reducing interoperability and re-use of data-
sets and software. Bespoke formats are often incompatible
and require significant time and compute resources spent in
data wrangling, and generally reduce the amount of FAIR
data that is available to scientists. Without a formal body
to declare such specifications or dedicated funding to pro-
duce a single solution for users, work is left to the commu-
nity to discuss and implement with the available resources.
The larger the community consensus, the more tools can be
adapted with the agreed upon solution. In turn, the lives of
the users in their daily activities become easier. Our work
on OME-Zarr to date shows an example of how community
consensus and investment can achieve concrete progress
towards FAIR data formats and tools.
The initial work to support OME-Zarr focused on
plugins for the primary desktop visualization and analy-
sis platforms—napari and Fiji, as well as a web browser
viewer. Each new specification was implemented in these
1
https:// zarr. dev/.
Histochemistry and Cell Biology
1 3
applications in order to prevent bias towards a single plat-
form. This was the state of the ecosystem for the initial
release at the end of 2021: functional with substantial lan-
guage support, but insufficient adoption to consider the for-
mat mature.
In the intervening year, the number of released tools that
work with OME-Zarr has increased significantly and the
amount of data available is growing similarly. This trend is
also visible in domains outside of bioimaging with institutes
like NASA preparing for releases of their data in Zarr-based
formats as part of their “Year of Open Science”
2
(Durbin
etal. 2020), (Ramachandran etal. 2021). The NGFF com-
munity finds itself in a very exciting phase. There is now a
cloud-optimized, chunked format that functions as a com-
mon API for both desktop, cluster, and web-based tools as
well as national and international repositories. Institutes and
repositories are working towards publishing their data in a
common format. For users, this means that many of their
most common scalability issues can be addressed by a solu-
tion that currently exists.
At the highest level, an OME-Zarr is a single bioimaging
container for multiple image acquisitions as well as derived
data. The versatility of the format stems in part from the under-
lying data format, Zarr, and in part from the OME-NGFF
community-defined specifications that are encoded in the meta-
data of the Zarr format, enabling use-cases across bioimaging
domains. The development of Zarr features and new specifi-
cations is accelerating, but already they provide the features
necessary to remove roadblocks to daily work.
Big data
OME-Zarr has been designed for performant reading and
writing of large image data. This begins by storing the
arrays of data in individual N-dimensional chunks. Since
pixels that are shown together in viewers are stored together,
they can be loaded more quickly. In a lightsheet dataset,
for example, a 3-dimension region of 128 × 128 × 64 pixels
might be colocated in a single atomic object. The current
specification
3
supports up to 5 dimensional images (time
point, channel, z, y, x). In the forthcoming 0.5 specification,
this constraint will be relaxed to allow N-dimensional arrays.
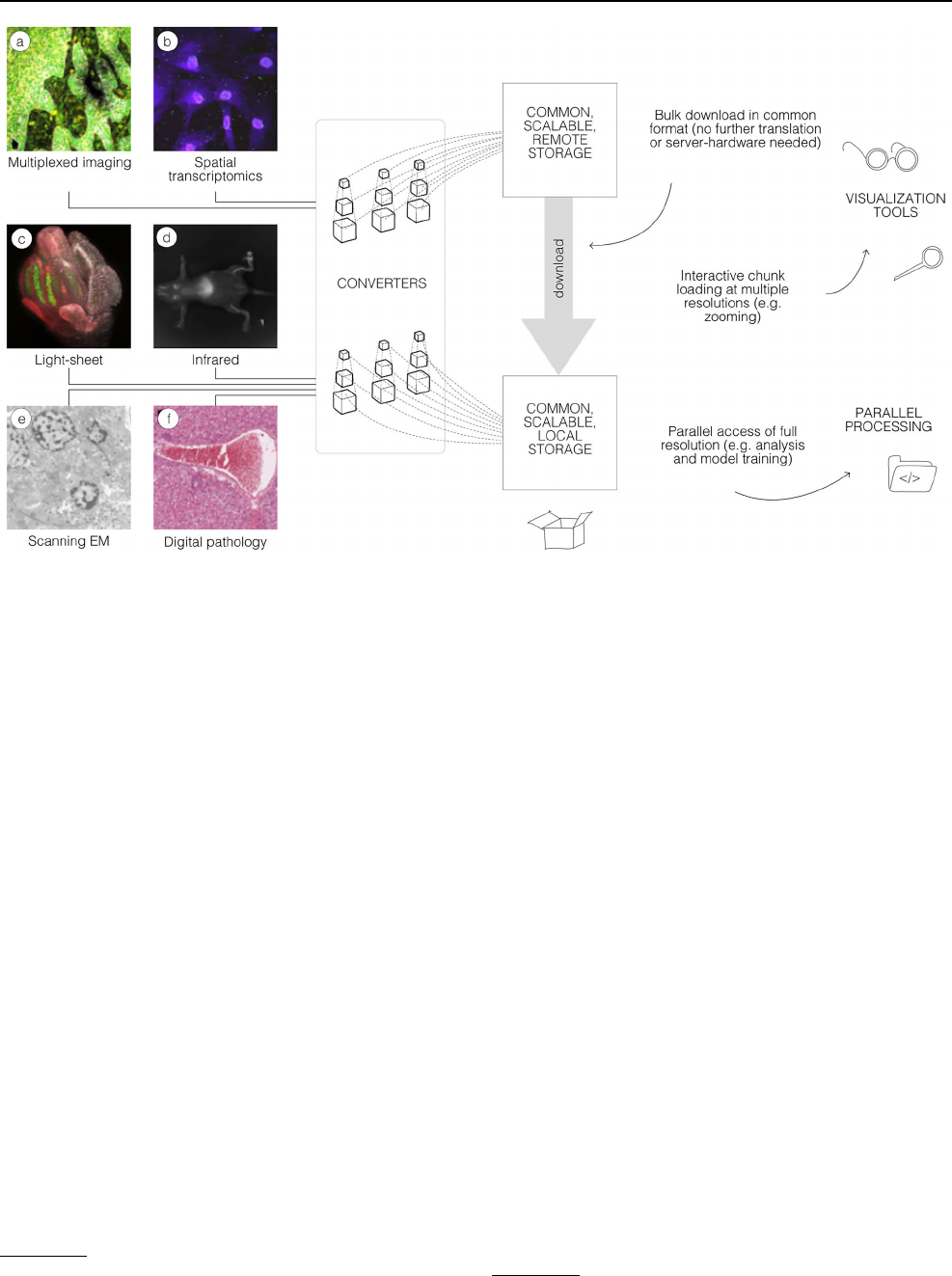
Fig. 1 A common format enables a diverse set of use cases via a
consistent API. A wide range of modalities can be converted into a
representation that can be equally accessed by a variety of tools.
This format can be used to download entire datasets for local pro-
cessing, to stream pyramidal sub-resolutions for interactive viewing
or to process entire resolutions in parallel. OME-Zarr data shown
includes a idr0076 (Ali et al. 2020), b idr0101 (Payne etal. 2021),
c idr0077 (Valuchova etal. 2020), d S-BIAD548 (Lim etal. 2023), e
S-BIAD217 (de Boer etal. 2020), and f S-BIAD501 (Igarashi etal.
2015)
2
https:// nasa. github. io/ Trans form- to- Open- Scien ce/ year- of- open-
scien ce/.
3
https:// ngff. openm icros copy. org/0.4.
Histochemistry and Cell Biology
1 3
To reduce file sizes and transfer times, Zarr supports com-
pression of each of the chunks. The compression algorithm
(e.g., GZIP or Blosc (Alted 2010)) and its parameters can
be configured transparently at the storage layer. The size of
chunks is configurable allowing users to choose the opti-
mal setting for a given use case to achieve a fine balancing
between file size, number of files, and overall read and write
speed for specific access patterns.
To allow smooth Google Maps-style zooming into large
images, OME-Zarr supports storage of image chunks at mul-
tiple resolution levels. Viewers can load data at the appropri-
ate resolution level (i.e., level of detail), which enables effi-
cient access to data even from remote storage. Furthermore,
many processing steps can be executed more efficiently on
smaller representations of the data.
Transparent organization
Another key characteristic of OME-Zarr is the ability to
organize multiple such multi-dimensional pyramids into a
hierarchy and attach metadata at each level of that hierarchy.
This is achieved with Zarr “groups” which contain Zarr
arrays and other groups in a hierarchical fashion. Meta-
data can be attached to each group and array separately in
web-readable JSON files. These features of the Zarr format
enable storing related data together, maintaining provenance
information. For example, a raw image, its deconvolution,
and even its segmentation can all be grouped together with
the metadata defining a consistent interpretation of the data.
Moreover, the community can make use of this metadata
organization to flexibly store further metadata schemas.
Where in OME-TIFF files, a single location is provided for
storing OME-XML, OME-Zarr makes possible the storage
of multiple standards such as “Recommended Metadata for
Biological Images”, REMBI (Sarkans etal. 2021), “Mini-
mum information guidelines for highly multiplexed tissue
images”, MITI (Schapiro etal. 2022), or “Quality Assess-
ment and Reproducibility for Instruments & Images in Light
Microscopy”, QUAREP-LiMi (Nelson etal. 2021) alongside
the OME-XML metadata.
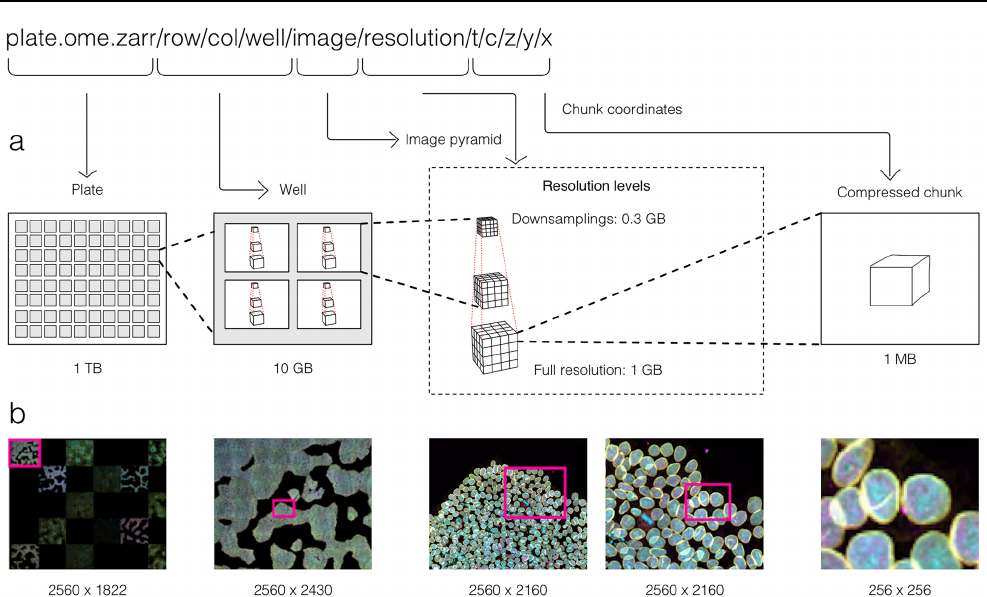
Fig. 2 By making use of an annotated hierarchy of arrays, OME-Zarr
can represent complex relationships between images, capture the
multiple resolutions of an image pyramid, and provide tunable chunk
size and compression all within a single abstraction layer that can
be saved as a directory of files on disk or shared remotely. a Each
level of nested directories provides a different level of abstraction:
the top-level directory can represent an entire 1 Terabyte plate with
more than 100,000 pixels in the X and Y dimensions, while the low-
est level directory represents individual chunks of N-dimensional data
as small as 1 Megabyte. b In the example shown, a concatenation of
low-resolution images produces a 2560 pixels × 1822 pixel represen-
tation of the entire plate, followed by similar examples of how many
pixels must be loaded by a client at each zoom level

Histochemistry and Cell Biology
1 3
Collections ofimages
With these combined capabilities, complex, data-rich col-
lections can be constructed to support diverse applications.
The OME-NGFF specification for high-content screening
(HCS)
4
, for example (Fig.2), defines multiple levels of hier-
archy for storing a plate, each of its wells and each of the
fields of that well as a separate image pyramid. Similarly,
segmentations, known in other domains as “annotations”,
“regions-of-interest” or “segmentation maps”, can be stored
as labeled images beside the raw image that was segmented.
Multi-modal datasets that represent multiple acquisitions on
the same sample can be stored along with location informa-
tion so that the images can be overlaid on one another for
visualization without changing the original underlying data.
Next steps
OME-NGFF specifications are being regularly proposed,
discussed, and implemented by the community to more
accurately capture bioimaging data. For example, a specifi-
cation for tables that annotate label images is slated for the
upcoming 0.5 version. Based on the heavily-used AnnData
table representation (Virshup etal. 2021), the objective of
the label table specification is to store measurements and
annotations on instances in a label image, including numeric
values, categorical values (e.g., strings), sparse arrays (e.g.,
touch matrices to encode neighborhood relationships), and
dense arrays (e.g., centroid coordinates). An early proto-
type of this idea from HuBMAP visualizes CODEX imaging
and segmentation data in combination with segmentation
centroids and clustering results simultaneously with the
Vitessce framework
5
(Keller etal. 2021). Other specifica-
tions currently under consideration include more complex
transformations to support correlative microscopy, efficient
point cloud storage, and the versioning of data and metadata
changes.
Another key next step will be how to support the NGFF
model in other storage scenarios. Being based originally on
the HDF5, Zarr’s compatible feature set makes the model
readily transferable between the two. This would provide
the user complementary approaches for balancing scalabil-
ity versus complexity. On the one hand, while the internal
structure of monolithic files like HDF5 are often described
by complex binary data structures accessible via libraries,
each Zarr chunk can be referenced via predefined, externally
stable paths which provide direct access of all chunk data and
metadata at each hierarchy level and can be listed by stand-
ard file browsers. With many storage backends, this strategy
enables the parallel writing of large image datasets, essential
for cluster and cloud-based processing. On the other hand,
the potentially large number of files produced by Zarr can
create problems on some file systems, generally increasing
the time to list, copy, and delete data. Having support for both
gives users a choice while the use of a common model in both
formats increases overall interoperability.
This and future strategies for meeting user requirements
will need periodic review. An upcoming version of Zarr, v3,
will support a sharded layout which places a configurable
number of chunks into a single shard file. This reduces the
total number of files at the cost of some writing parallelism.
A similar feature is available in HDF5 using “external” files
and “virtual datasets” to group many separate files together.
Users looking for the optimal solution will need to carefully
consider the trade-offs, e.g., the impact of a multi-file for-
mat on the average consumer while existing tools are being
updated.
Selection ofOME‑Zarr Tools
Many common difficulties in image handling and analy-
sis stem from both a lack of consistency and compatibility
between data inputs and outputs and the resulting siloization
of available tools. Without assistance, software packages are
often only able to ensure compatibility with a small portion
of formats. A common strategy to deal with the proliferation
of file formats is to translate from one of the many current file
formats on the fly. This is how open-source libraries like Bio-
Formats (Linkert etal. 2010) provide access to applications
as diverse as Fiji and OMERO. Translation can contribute
significantly to the scalability challenge. Additionally, meta-
data can get lost during image translation due to opaque file
structures, leaving users to provide most metadata when shar-
ing or submitting to public resources. Sharing and re-use is
complicated by disconnected images. Minimizing the number
of file formats and standardizing the included metadata, in
turn, fosters collaboration and stability.
The original release and publication of the OME-Zarr
format was accompanied by three tools—one in Java, one in
Python, and one in the web browser—that could be used to
visualize and work with OME-Zarr data (Fig.3). Over the
course of the subsequent year, the number of tools has grown
significantly covering additional use cases. Several of these
applications originally developed their own custom format
internally in order to achieve the performance they needed
but have now added support for OME-Zarr allowing them
to interoperate with one another.
Below we provide an updated list of tools that were
known to handle OME-Zarr at the time of writing. This
list, however, will quickly age post-publication. In order
to keep track of the software packages which have added
4
https:// ngff. openm icros copy. org/0. 4/# hcs- layout
5
https:// vites sce. io.

Histochemistry and Cell Biology
1 3
support for OME-Zarr, a registry has been created at
https:// ngff. openm icros cop y. org/ tools. Our list is catego-
rized into three large, though at times overlapping, cate-
gories. We start with the visualization tools (Table1) that
are broadly useful for interactively working with data.
They provide an overview of what is possible with OME-
Zarr. Where possible links to example data have been
provided. A list of libraries follows (Table2) that can
be used to automate operations on OME-Zarr. These are
useful especially when building pipelines or automating
a workflow. Finally, generators (Table4) are used to take
data either from other tools or from the original acquisi-
tion system and create OME-Zarr data.
Visualization
AGAVE
AGAVE
6
(Fig.4) is an open-source native application for
high quality GPU rendering of multichannel volume data.
It uses a photorealistic path tracing algorithm to produce
images with lighting and shadows, allowing greater detail
and higher interpretability of spatial relationships within the
data. AGAVE is implemented in C++ with OpenGL and
runs on Windows, MacOS and Linux. OME-Zarr support
is implemented through the TensorStore library, described
below. AGAVE provides a memory estimate and allows
selection of the multiresolution level and slice ranges in
the XYZ dimensions. Future work in AGAVE will include
the ability to combine OME-Zarr data from multiple data
sources and improvements for more quantitative viewing
such as display of physical units, voxel intensities, and a 2D
slice display mode.
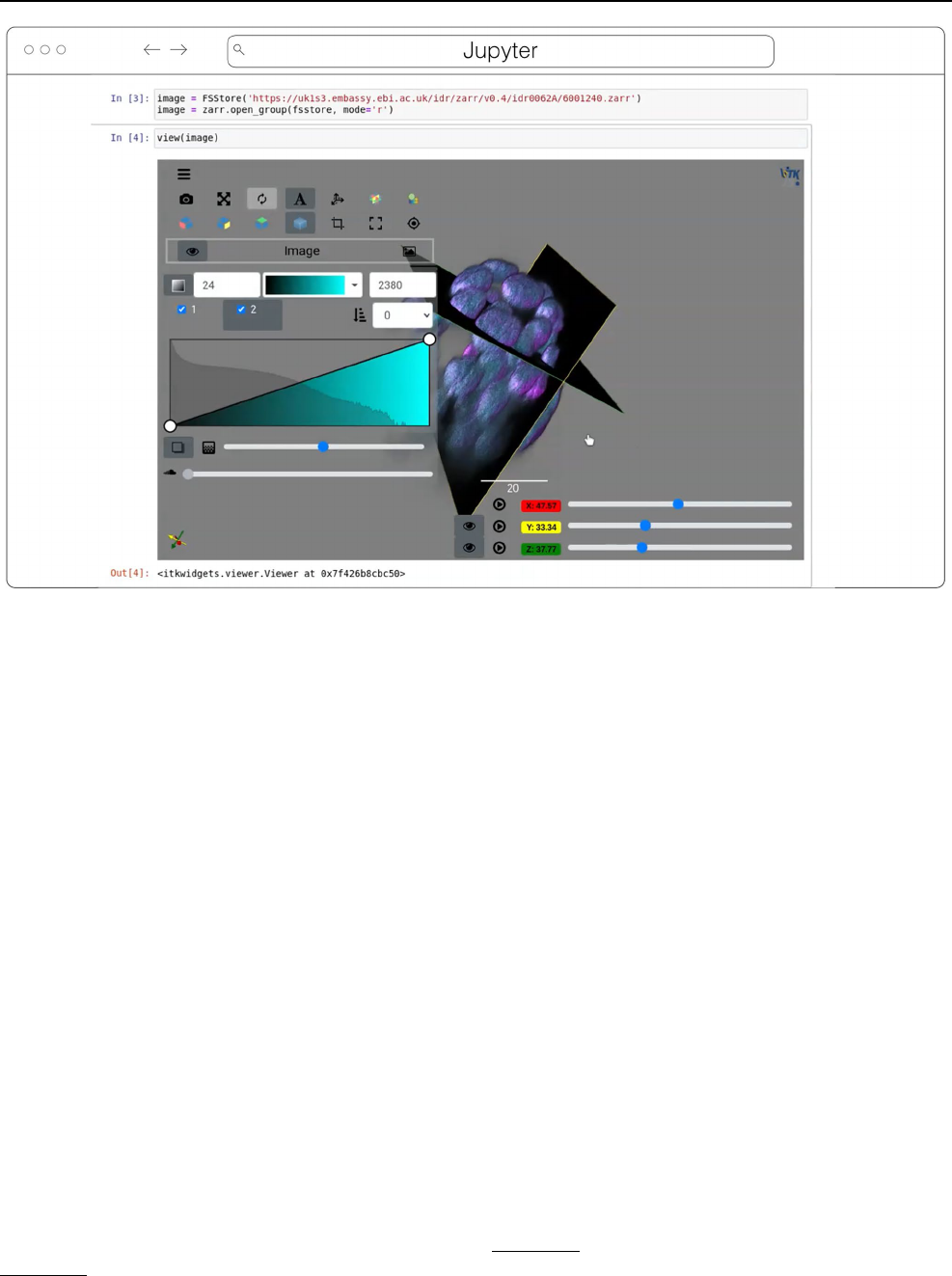
ITKWidgets
ITKWidgets (Fig.5) provides interactive widgets to visu-
alize images, point sets, and 3D geometry on the web
(McCormick etal. 2022). The development of ITKWidg-
ets was motivated by the need for interactive insights into
N-dimensional scientific datasets, such as three-dimensional,
multi-channel bioimages. ITKWidgets is a component of the
Fig. 3 The original viewers of OME-Zarr published in Moore etal.
2021, from left to right BigDataViewer, napari, and Vizarr, here seen
loading a view of the same EM volume of a 6 day old Platynereis
larva from (Vergara et al. 2020) available at https:// s3. embl. de/ i2k-
2020/ platy- raw. ome. zarr. These three applications provided broad
coverage over the most common bioimaging platforms like Fiji and
napari but critically also a web viewer that could stream data on the
fly
Table 1 List of visualization
tools in the order they are
described below along with
their primary platform of use
and the software frameworks
used to build them
An up-to-date version of the table is maintained at https:// ngff. openm icros copy. org/ tools and contributions
are welcome
Visualization tool Use Language/framework
AGAVE Linux, MacOS, Windows C++, OpenGL
ITKWidgets Web (Jupyter) Python, WASM
MoBIE/BigDataViewer Linux, MacOS, Windows Java
napari Desktop Python
Neuroglancer Web WebGL
Validator Web Svelte
Viv Web React, deck.gl
webKnossos Web React, WebGL
website-3d-cell-viewer Web React, TypeScript, WebGL
6
https:// www. allen cell. org/ patht race- rende ring. html.
Histochemistry and Cell Biology
1 3
Insight Toolkit (ITK), an open-source, cross-platform suite
of libraries and tools for N-dimensional spatial analysis and
visualization (McCormick etal. 2014).
Designed for web-first visualization and large-scale data,
ITKWidgets is built on universally deployable technologies
and the OME-NGFF and ITK data models. ITKWidgets com-
municates with Google CoLab, Jupyter Notebooks, Jupyter-
Lab, and JupyterLite with ImJoy, a modern remote procedure
communication library and plugin interface for biomedical
computing in the deep learning era (Ouyang etal. 2019).
ITKWidgets is available as a Python package or the client-
side viewer application can be loaded by visiting its webpage.
In Python, NumPy, PyImageJ, PyTorch, the Visualization
Toolkit (VTK), ITK, Xarray, Zarr, and Dask data structures
are transformed on-demand to multiscale OME-Zarr. In the
browser, ITKWasm will generate a multiscale OME-Zarr on-
demand from common bioimaging file formats (McCormick
2022). In Python, a simple view command accepts datasets
for interactive exploration. In the client-side application, a
local dataset can be selected or a shareable URL with the
dataset and rendering parameters can be generated.
With a focus on supporting registration (alignment), ITK-
Widgets is recommended for the comparison of datasets. Spa-
tial metadata on multi-dimensional raster images along with
associated point-based volumetric data, geometries, and anno-
tations are supported to understand their relationship in space.
Additionally, this provides a foundation for the creation of spa-
tial transformations that define or improve on the alignment
of datasets. ITKWidgets is particularly focused on providing
elegant renderings to elucidate insights from volumetric infor-
mation. Advanced rendering capabilities, such as light scatter-
ing, are supported. Intuitive and efficient interactive widgets
are available to select rendering parameters, such as color maps
and opacity transfer functions. The rendering system leverages
OME-Zarr chunked, multiscale architecture to automatically
load the optimal amount of data for a selected volumetric region
by accounting for the current system's hardware capabilities.
The user interface is customizable via vanilla HTML/
CSS/JavaScript or web frameworks such as React.js or Vue.
js, and the ability to present simplified versions of current
interfaces and transparently integrate the viewer into larger
applications is improving. This flexibility enables integra-
tions into custom applications such as TensorBoardPlug-
in3D, a plugin for TensorBoard to support the development
of deep learning models on 3D images (Major and McCor-
mick 2022). Scalability will be achieved through bolstered
OME-Zarr data model support.
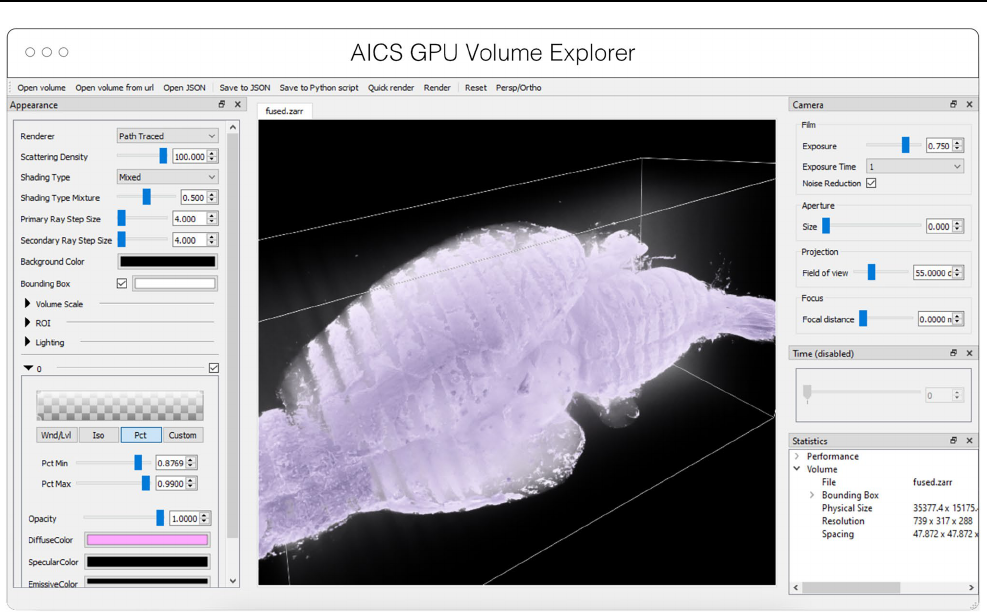
Fig. 4 Advanced GPU Accelerated Volume Explorer (AGAVE) dis-
playing a downsampled level from a multi-terabyte mouse brain
OME-Zarr dataset. The number of pixels actually loaded is displayed
at lower right. The full resolution data is 47,310 × 20,344 × 18,471
which consumes about 33TB. The ability to quickly access multires-
olution data makes low latency interactive visualization possible
Histochemistry and Cell Biology
1 3
MoBIE/BigDataViewer
MoBIE
7
(Fig.3) is a Fiji plugin (Schindelin etal. 2012) for
the exploration of big, possibly remote, image datasets (Pape
etal. 2022). The development of MoBIE was initiated in 2018
at EMBL Heidelberg in order to solve the challenge of brows-
ing and publicly sharing a large CLEM dataset consisting of
one high-resolution TB sized 3D volume EM dataset, cell and
tissue segmentations of the EM data, tables with segmentation
annotations, and around 200 registered lower resolution LM
images (Vergara etal. 2020) and is still in daily use across the
institute.
The main usage of MoBIE is to continuously browse
complex image datasets from the moment they are produced
up until publication. A typical workflow is to use other appli-
cations for image and data analysis and add the output of
those applications such as segmentations and tables into the
corresponding MoBIE project for visual inspection, explo-
ration and quality control. An exception is the possibility
to perform semi-manual image registration directly within
MoBIE by means of an integration with the BigWarp Fiji
plugin (Bogovic etal. 2016).
MoBIE is a desktop application written in Java that
heavily relies on BigDataViewer
8
(Pietzsch etal. 2015) for
image rendering and the N5 library
9
for (remote) image I/O,
described below. It supports viewing locally (e.g. file-system)
and remotely (e.g. “Simple Storage Service”, or S3) hosted
OME-Zarr image data as well as HDF5 and the eponymous
N5 multi-scale image data format. In addition to simply
viewing OME-Zarr images in Fiji, the main usage and fea-
ture of MoBIE is the ability to structure potentially thousands
of images into a “MoBIE project” and define and configure
useful views into that dataset. An important application of
those features are “MoBIE views” that can be configured to
conveniently browse the raw data associated with figures in
publications.
In the future, MoBIE will support interactive deep-learn-
ing based image segmentation by means of an integration
with the BioImage Model Zoo (Ouyang etal. 2022). It will
also be shipped as a conda package for opening images,
segmentations and tables from the command line. This will
support the visual inspection of the output of image segmen-
tation and feature extraction algorithms. Another planned
Fig. 5 ITKWidgets 3D rendering an OME-Zarr for IDR 0062A in Jupyter. Interactive features shown include volume rendering, slicing planes,
and interactive widgets to adjust rendering parameters and slice planes indices
7
https:// mobie. github. io/.
8
https:// github. com/ bigda tavie wer.
9
https:// github. com/ saalf eldlab/ n5.

Histochemistry and Cell Biology
1 3
feature is the rendering of the HCS specification of OME-
Zarr as a plate layout.
napari
napari
10
(Fig.3) is a multi-dimensional data viewer written
in Python (Sofroniew etal. 2022). Many different types of
data can be viewed and annotated in napari including multi-
dimensional images, point clouds, polygons, tracks, and
meshes. napari can be scripted using its Python API, used
interactively via interactive environments such as IPython
(Perez and Granger 2007) and Jupyter notebooks (Granger
and Pérez 2021), and launched from the command line.
While the core napari package is focused on interactively
viewing and annotating data, it can be extended to other
use cases via custom scripts or through the plugin interface.
OME-Zarr data can be viewed in napari via the napari-
ome-zarr plugin
11
. Users can load OME-Zarr datasets through
the command line interface or via the Python API. Datasets
can be loaded from both local and remote data sources. Local
OME-Zarr files can also be loaded via drag & drop. Develop-
ers can use the ome-zarr-py library to load datasets and add
them to the viewer via the Python API. The Fractal framework
uses Dask lazy loading with the napari-ome-zarr plugin and
the experimental napari asynchronous loading feature (under
development, NAP-4
12
) to interactively view 2D multichan-
nel datasets from 100s of GBs to 1TB in size (see Fractal
section below). The SpatialData framework (Marconato etal.
2023) also combines Dask lazy loading and the napari plugin
napari-spatialdata
13
to visualize spatial omics data, that often
entails a variety of data types: raster images, points, polygons
and annotations.
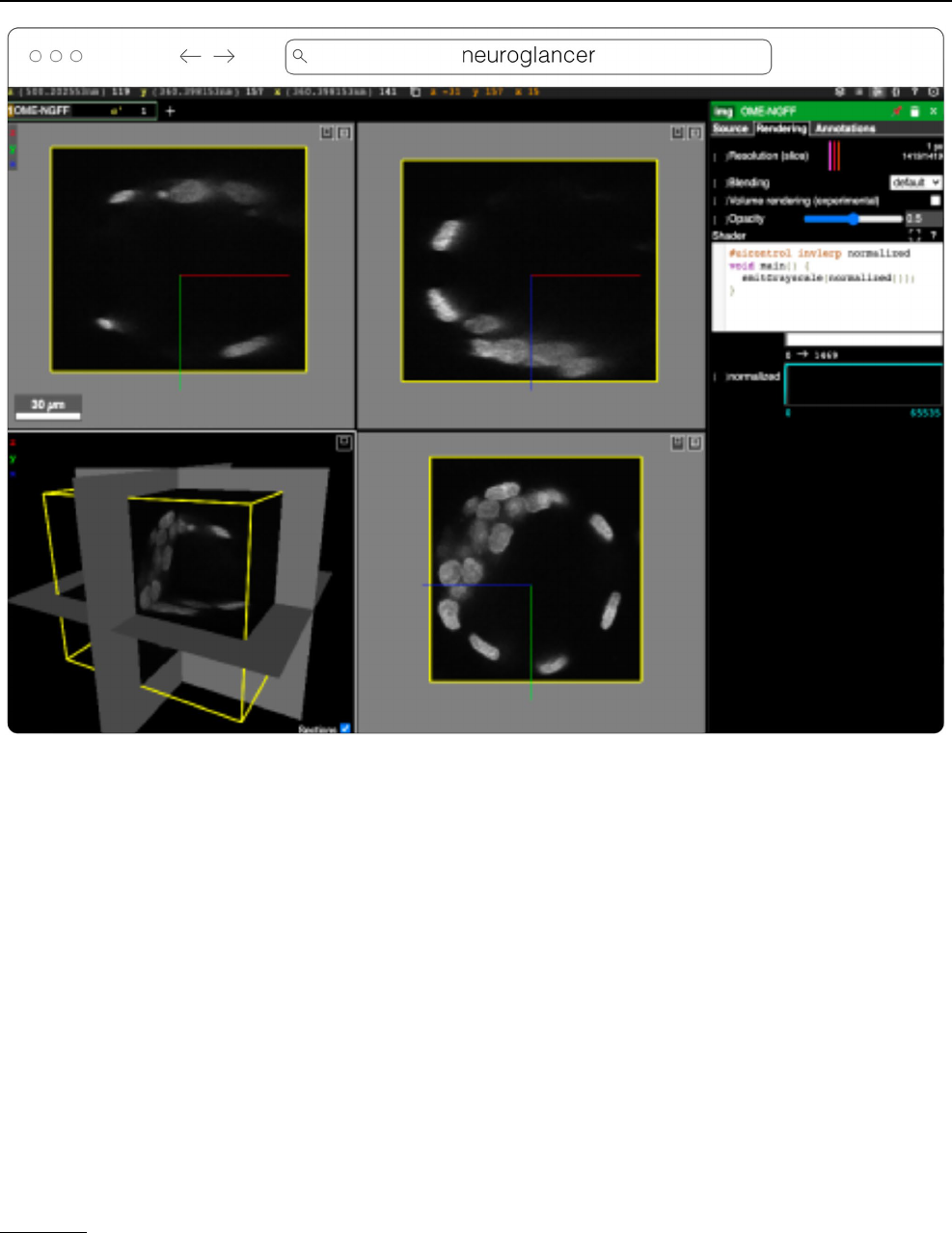
Neuroglancer
Neuroglancer
14
(Fig.6) is an open-source web-based visuali-
zation tool for multi-dimensional volumetric data. Originally
designed for visualizing petabyte-scale volume electron
microscopy datasets of brain ultrastructure, it is now widely
used to visualize scientific imaging data in many different
application areas, including connectomics, lightsheet func-
tional calcium neuroimaging, fMRI, and high-throughput
screening. Key functionality includes:
•
Scalability to petabyte and larger datasets through the
use of multi-resolution data streaming for OME-Zarr and
other chunked formats
•
Cross-section views at arbitrary oblique angles
•
Rendering of segmentations and meshes
•
Arbitrarily many datasets may be displayed tiled side-by-
side, or overlaid as separate “layers”
•
Mapping from stored data to displayed RGBA values
may be customized through user-defined "shader" func-
tions that can be edited directly within the tool, and these
shaders can make use of user-defined UI controls such as
sliders and checkboxes
•
Experimental volume rendering support
•
Supports Zarr data served from arbitrary HTTP servers,
as well as Google Cloud Storage (GCS) and Amazon S3
Neuroglancer is built using WebGL and relies on advanced
GPU rendering and compression techniques to achieve high
performance despite the limitations of the web platform. As
a web-based tool, Neuroglancer is particularly convenient for
collaborating on datasets; users can share particular views of
a dataset simply by sharing a URL. As a purely client-side
web application, Neuroglancer can be used directly from the
official hosted instance, or it can be deployed to any static file
web server. There is also a Python package (`neuroglancer` on
PyPI) that allows for full interaction with the viewer state from
Python, defining of custom key and mouse bindings that invoke
Python callbacks, and also allows Neuroglancer to display in-
memory NumPy arrays, as well as arrays from other packages
such as TensorStore, zarr-python, Dask and h5py that provide a
similar NumPy-like interface. The Python package can be used
both in standalone Python programs and shells and also from
Jupyter notebooks, and provides a convenient way to quickly
build ad-hoc data analysis and proofreading tools.
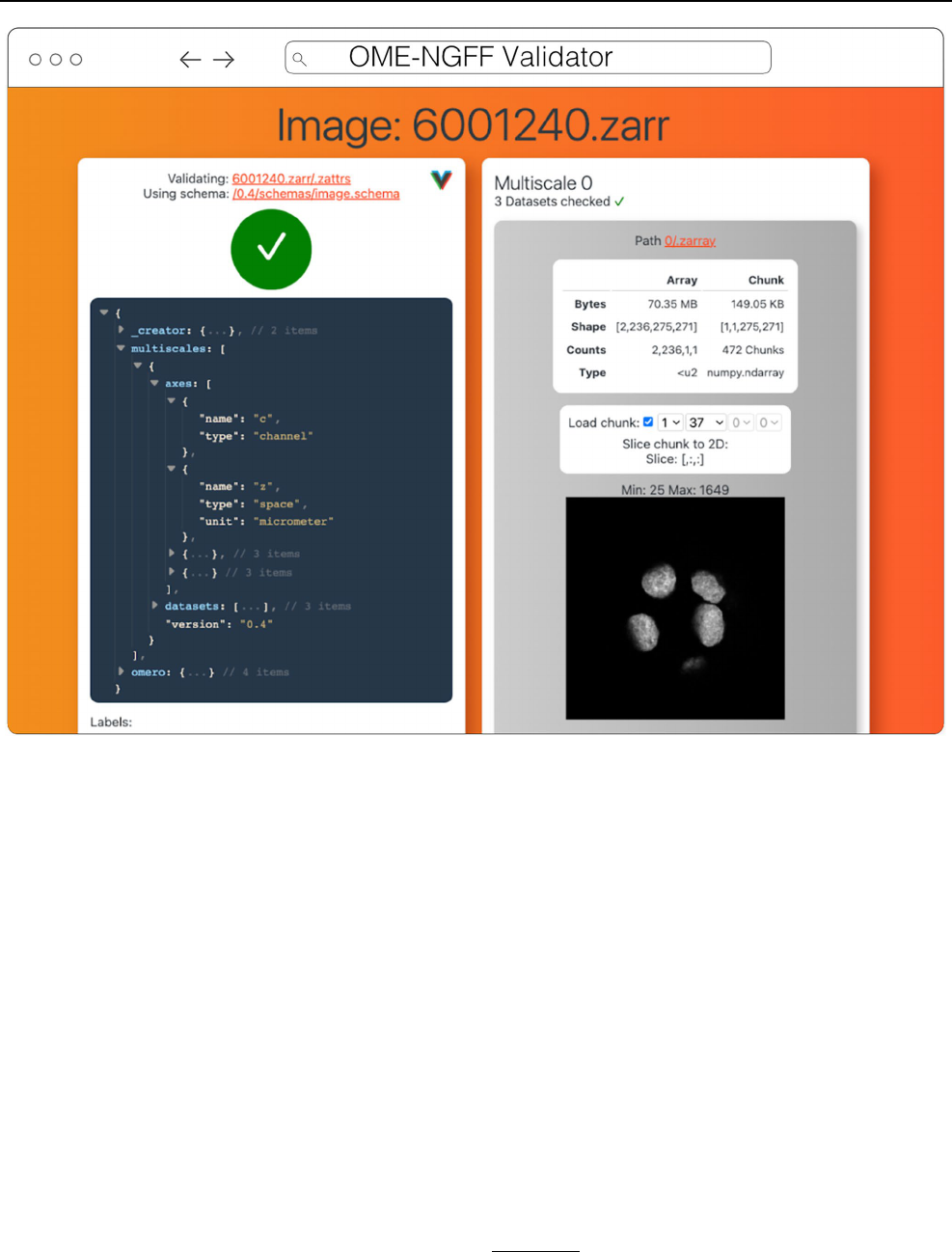
Validator
The ome-ngff-validator
15
(Fig.7) is a web-based tool for val-
idating and inspecting OME-Zarr data. It uses schema files
(in the JSON schema format) for validating the JSON data of
OME-Zarr, and uses zarr.js for loading image data chunks.
Providing the community with an easy way to validate their
data is an important part of promoting the adoption of OME-
Zarr. When newly developed tools are exchanging data in
this format, it is essential to know whether the data complies
with the OME-Zarr specification. It is also useful to be able
to browse and inspect the data in order to troubleshoot any
issues with creation or reading of the format.
A web-based tool is convenient for users as they do not
need to install any software, and it also means that they can
10
https:// napari. org/.
11
https:// github. com/ ome/ napari- ome- zarr.
12
https:// napari. org/ stable/ naps/4- async- slici ng. html.
13
https:// github. com/ scver se/ napari- spati aldata.
14
https:// github. com/ google/ neuro glanc er.
15
https:// github. com/ ome/ ome- ngff- valid ator.
Histochemistry and Cell Biology
1 3
share a URL that shows their data in the ome-ngff-valida-
tor
16
. This improves the ability of the community to discuss
and work with public OME-Zarr files.
Viv
Viv
17
is an open-source library for WebGL-based 2D mul-
tiscale, multi-channel, image rendering with the ability to
handle 3D fixed-resolution volumes as well (Manz etal.
2022). Separation of rendering and data-loading implemen-
tations in Viv allows the library to provide functionality for
fetching data directly from multiple open-standard data for-
mats. OME-TIFF and OME-Zarr images can be loaded using
Viv and directly rendered on the GPU. Viv is built using
deck.gl, a high-level API for WebGL that simplifies building
interactive spatial (i.e., Cartesian coordinate-based) and geo-
spatial data visualizations. In deck.gl parlance, Viv simply
provides “layers” that can be composed with other layers
or operations to create complex visualizations. As a result,
developers using Viv can take advantage of the wider eco-
system around deck.gl that includes custom layers, modes
of interactivity, mathematical operations, and shaders. This
flexibility was core to the design of the Viv API, as end-
users can extend the provided layers or define custom data
loaders.
Viv was initially motivated by the need of members of
the HuBMAP consortium to display high resolution, multi-
modal (i.e., overlaid) images within the HuBMAP data por-
tal (e.g., http:// vites sce. io/#? datas et= neuma nn- 2020). Work-
ing within the constraints of limited engineering resources,
the data portal development team aimed to avoid running
and maintaining server-side pre-rendering infrastructure.
Further, FAIR data access principles were paramount to the
creation of the data portal. The development of Viv enabled
Fig. 6 Neuroglancer rendering the same OME-Zarr from IDR 0062A as Fig.5
16
https:// ome. github. io/ ome- ngff- valid ator/? source= https:// uk1s3.
embas sy. ebi. ac. uk/ idr/ zarr/ v0.4/ idr00 62A/ 60012 40. zarr.
17
https:// github. com/ hms- dbmi/ viv.
Histochemistry and Cell Biology
1 3
rendering images within a web page, thereby enabling the
data portal server-side infrastructure to be as simple as a
static file server or a cloud object storage system. Adoption
of FAIR data access principles in the consortium motivated
the implementation of data loaders for open-standard for-
mats. Viv has been adopted by other consortia including the
Kidney Precision Medicine Project (KPMP) (de Boer etal.
2021) as well as the Galaxy Project (Galaxy Community
2022) to address data sharing and visualization challenges.
Wellcome Sanger Institute in collaboration with Newcastle
University is working on a human whole embryo project that
leverages different spatial technologies to create a holistic
view of human embryo at the single cell level. Vitessce,
which is built on top of Viv, is used for visualizing both the
single cell sequencing and imaging data simultaneously that
are saved as OME-Zarr.
In addition to the bioimaging rendering challenges that
Viv addresses in production, it has also served as a testbed
and mechanism to prototype new data standards. Vizarr
18
(Fig.3) is a bioimaging visualization tool built using Viv
that served as one of the first implementations of a reader
and renderer for the HCS metadata standard in OME-Zarr
19
.
Using Viv as the core rendering library, Vizarr is able to
simultaneously render hundreds of images from an HCS
dataset. The design of Viv as a UI-agnostic library that
separates rendering from data loading means that it will
remain possible to quickly develop or adapt existing appli-
cations to the evolving and increasingly flexible OME-NGFF
specification.
Fig. 7 OME-NGFF Validator validating the same image from IDR 0062A on the left, and on the right providing a summary of the size of the
data as well as providing a quick visualization of a single plane
18
https:// github. com/ hms- dbmi/ vizarr.
19
https:// www. openm icros copy. org/ 2020/ 12/ 01/ zarr- hcs. html.
Histochemistry and Cell Biology
1 3
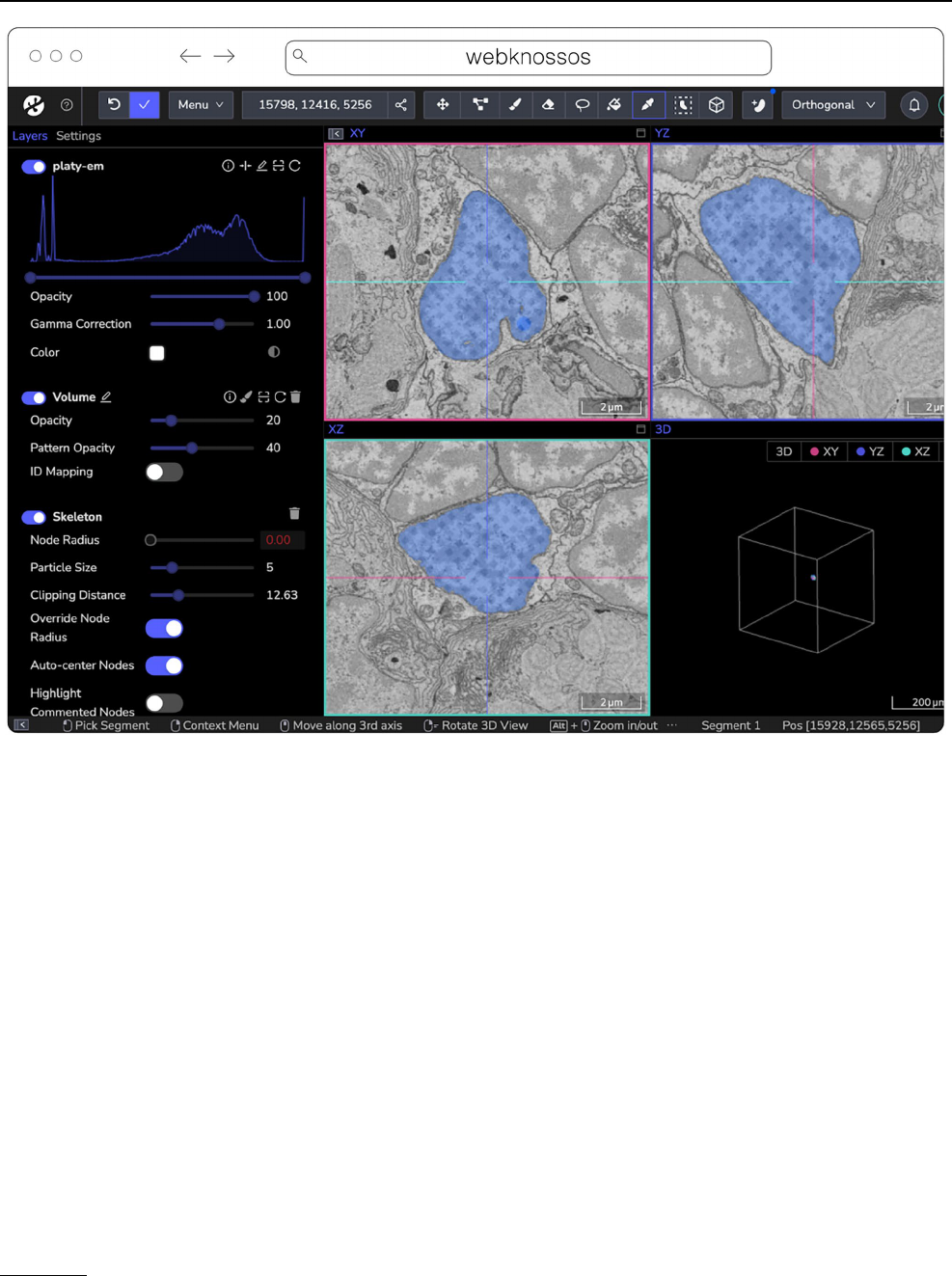
webKnossos
webKnossos
20
(Fig.8) is a web-based, open-source platform
to visualize, share and annotate very large image datasets
collaboratively (Boergens etal. 2017). webKnossos was
originally developed to distribute 3D image datasets of a
few hundred gigabytes to students for annotation. It fea-
tures segmentation tools to generate label masks for manual
analysis or as training data for deep learning-based analysis.
Combined with mesh rendering features, webKnossos can
visualize large-scale segmentations. Additionally, webKnos-
sos features efficient skeleton annotation tools for creating
neuron traces. In many labs, webKnossos has become the
primary hub to store all image datasets in a common place
for lab members and external collaborators to access.
webKnossos works best for very large 3-dimensional
image data, such as EM, MicroCT and fluorescence. It
facilitates collaboratively annotating and visualizing data.
Users can:
•
visualize very large 3D datasets,
•
manually segment or skeleton data efficiently,
•
proof-read automatic segmentation,
•
share data with collaborators,
•
manage data in one place with access restrictions, and
•
stream data through an OME-Zarr-based API or use a
Python library to enable interoperation with other tools.
webKnossos allows users to store all data in one place
and access it from wherever you are, no matter the size of
the data, like a Google Drive for large microscopy images.
The server component stores all the image and annotation
data as well as user settings. In addition to server-stored
image data, remote datasets stored in OME-Zarr can also
Fig. 8 webknossos loading an EM volume of a 6day old Platynereis larva from (Vergara etal. 2020) with a manually added segmentation. The
web accessible version is accessible at https:// wklink. org/ 6422
20
https:// webkn ossos. org/.

Histochemistry and Cell Biology
1 3
be accessed from HTTP, S3 and GCS sources. webKnossos
uses GPU rendering for high-performance visualization of
the data. Users can access webKnossos through a browser
without the need for additional installations. The easiest
way to use webKnossos is to open an account on webknos-
sos.org. It is free for limited storage. Alternatively, both a
Docker-based setup and commercial private hosting ser-
vices are available.
webKnossos is a mature software and in routine use for
more than 10years. It gained support for OME-Zarr in 2022
and implements many features of the format. OME-Zarr
datasets can be imported via URL, optionally with creden-
tials. OME-Zarr data is read on the backend with on-the-fly
re-chunking and additional webKnossos-managed access
controls. Additionally, all data in webKnossos is accessible
as OME-Zarr data to be used in other tools via token-pro-
tected dynamic URLs. Therefore, the software can be used
as a central hub for teams to manage OME-Zarr datasets.
webKnossos is actively developed by a dedicated develop-
ment team with monthly releases. Today, most users are
from the Volume EM community, especially EM connec-
tomics and cell biology (Rzepka etal. 2023). However, light
microscopy users are also well-represented. In upcoming
versions, webKnossos will add support for image transfor-
mations, multi-modal datasets, and time-series data as well
as the ability to run AI-based segmentations and to show
segment statistics for quantitative analysis. Updated road-
map information is available under https:// webkn ossos. org/
roadm ap.
Website‑3d‑cell‑viewer (and volume‑viewer) fromAICS
The Allen Institute for Cell Science is an open science
organization, interested in producing results, data, and code
that can be shared with the world. Volumetric microscopy
datasets are presented for interactive exploration through the
Cell Feature Explorer
21
, a web-based application. As with so
many microscopy efforts, ever larger and larger datasets are
the norm. As an early part of transitioning storage and pro-
cessing to the cloud, the interactive 3D viewer component
of Cell Feature Explorer has been extended to read OME-
Zarr data.
The core of the viewer component is called volume-
viewer which implements all of the data loading and 3D
rendering, using WebGL and Typescript (Fig.9). The front
end, called website-3d-cell-viewer, is a reusable React com-
ponent that has also been published as a standalone viewer
22
and embedded in a Jupyter notebook plugin
23
.
This viewer is optimized for multichannel data and works
well with volumetric data where the xy resolution is higher
than z. The standalone version of the viewer supports OME-
Zarr through a URL query parameter. The OME-Zarr sup-
port is implemented using the Zarr.js library
24
. In its first
implementation, only TCZYX data is supported, only from
publicly accessible HTTP(S) URLs, and as of this writ-
ing only loads the lowest multiresolution level for memory
constraints. Coming enhancements include more general
data loading as well as user interface for selection of mul-
tiresolution level and time. There is also work being done to
be able to produce shareable URLs that capture all viewer
settings.
Beyond
More analysis tools leveraging the libraries to create derived
datasets and produce quantitative insights into OME-Zarr
data are already planned. For example, the BigStitcher (Hörl
etal. 2019) toolchain which currently relies on a custom
XML/HDF5 format will gain support for OME-Zarr, making
it easier to parallelize distributed access for HPC and avoid
additional file conversion. This is particularly important for
very large datasets and means that NGFF will be natively
exported by BDV in Fiji. Work on this is underway in the
bigdataviewer-n5
25
and bigstitcher-spark
26
repositories. At
the time of writing, BigStitcher has support for large file
export and limited direct support of OME-Zarr datasets
via the bigdataviewer-omezarr module
27
. This module pro-
vides a transient bridge between the current XML-based
dataset definition and OME-Zarr images until native sup-
port is implemented. It allows multiple images stored in one
NGFF dataset to be defined as separate view setups in BDV/
BigStitcher (Fig.10). Other tools such as RS-FISH (Bahry
etal. 2022) for spot detection and STIM for visualizing and
reconstruction of spatial transcriptomics datasets (Preibisch
etal. 2022) are currently being extended to support OME-
Zarr and other formats. Additionally, registration tools that
generate the developing spatial transformation specification
are planned to enable quantitative comparison of compatible
datasets.
To track these developments, the community will main-
tain https:// ngff. openm icros copy. org/ tools for finding the
status of other new and exciting developments.
21
https:// cfe. allen cell. org.
22
https:// allen- cell- anima ted. github. io/ websi te- 3d- cell- viewer/.
23
https:// pypi. org/ proje ct/ nbvv/.
24
https:// github. com/ gzuid hof/ zarr. js/.
25
https:// github. com/ mobie/ bigda tavie wer- n5.
26
https:// github. com/ Preib ischL ab/ BigSt itcher- Spark.
27
https:// github. com/ bigda tavie wer/ bigda tavie wer- omeza rr.
Histochemistry and Cell Biology
1 3
Libraries
Behind most of the visualization tools above and many
other applications are OME-Zarr capable libraries (Table2)
that can be used in a wide variety of situations. Workflow
systems like Nextflow or Snakemake can use them to read
or write OME-Zarr data, and the same is true of machine
learning pipelines like Tensorflow and PyTorch. Where
dedicated widgets like ITKWidgets are not available, these
libraries can make use of existing software stacks like Dask
and NumPy to visualize the data in Jupyter Notebooks or to
perform parallel analysis.
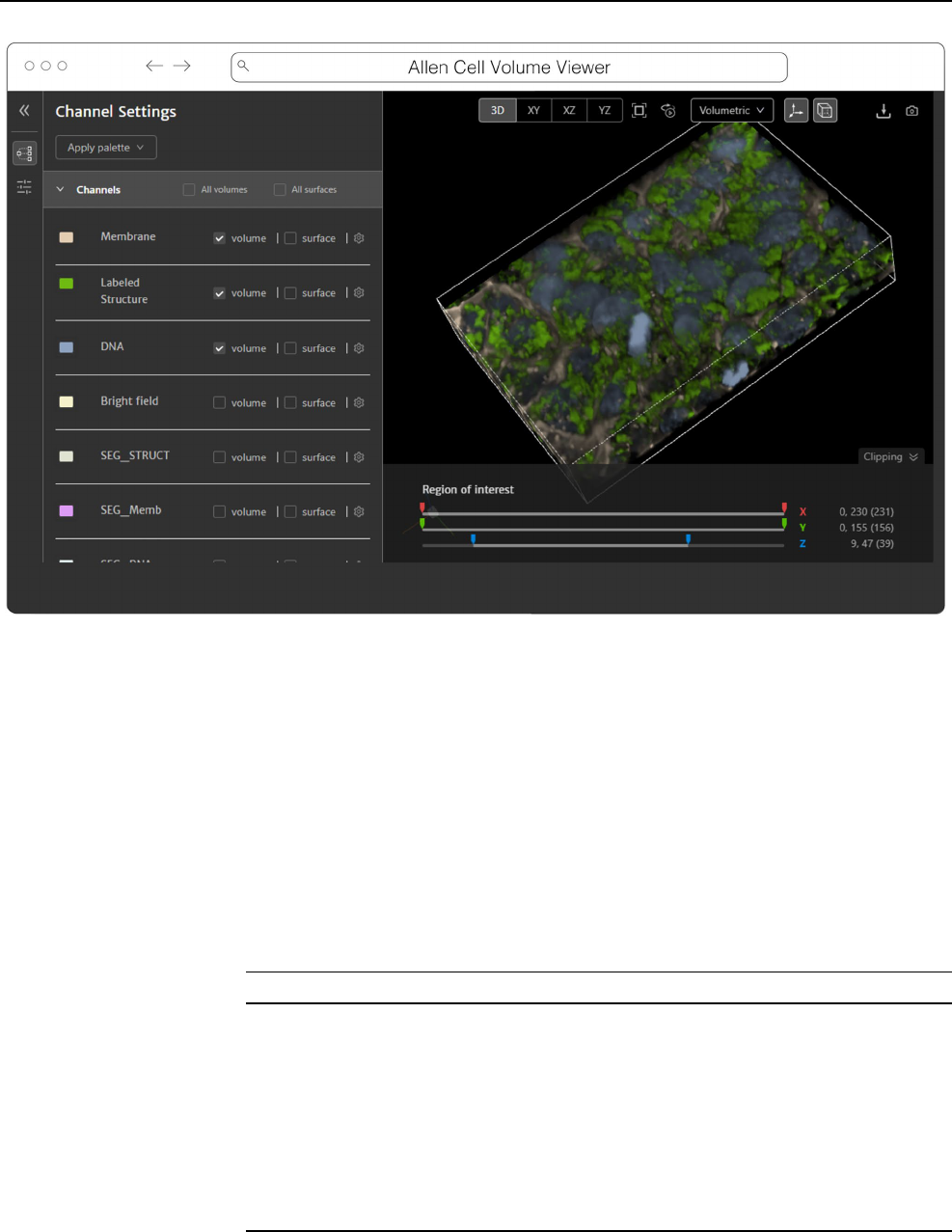
Fig. 9 Allen Cell Volume Viewer displaying a multichannel fluorescence image of gene edited hiPSC cells via a downsampled level of an OME-
Zarr converted from the dataset found at https:// cfe. allen cell. org/? datas et= varia nce_ v1
Table 2 List of libraries broken
down by programming language
in the order they are described
below along with a brief
description of their use
An up-to-date version of the table is maintained at https:// ngff. openm icros copy. org/ tools and contributions
are welcome
Language Library Comment
Python ome-zarr-py Reference implementation for NGFF specifications
AICSImageIO General bio data loading library
Fractal Framework for processing HCS images at scale
BFIO Optimized reading and writing of TIFF and Zarr
SpatialData Enable the alignment of spatial omics datasets
C++ TensorStore High-performance access to multiple array formats
Nyxus Out-of-core, parallel feature extraction library
Java
OMEZarrReader Plugin for reading OME-Zarr in Bio-Formats
N5 API Array data reading of Zarr, HDF5, and N5 formats
Histochemistry and Cell Biology
1 3
Python
ome-zarr-py
28
, available on PyPI, was the first implemen-
tation of OME-Zarr and is at the time of writing consid-
ered the reference implementation of the OME-NGFF data
model. Reading, writing, and validation of all specifications
are supported, without attempting to provide complete high-
level functionality for analysis. Instead, several libraries
have been built on top of ome-zarr-py. AICSImageIO is a
popular Python library for general 5D bio data loading. In
addition to loading OME-Zarr data, AICSImageIO provides
OmeZarrWriter using ome-zarr-py under the hood. In this
way, format conversion is possible by loading data with
AICSImageIO and immediately passing the Dask array to
OmeZarrWriter, though improvements in the metadata sup-
port are needed.
Fractal
29
is a framework to process high-content imag-
ing data at scale and prepare it for interactive visualization.
Fractal is focused on processing images in the OME-Zarr
format and saving results in forms of images, label images
and feature tables back into OME-Zarr, while keeping
orchestration of these processing steps cluster friendly via
slurm
30
. It allows users to build workflows to process images
in OME-Zarr files at the TB scale to segment images and
make measurements. As a result, large-scale OME-Zarr
image datasets can be processed by Fractal and then browsed
interactively across pyramid levels in viewers like napari
31
(Fig.2). Fractal is in its early stages and currently contains
workflows to be controlled from the command line interface.
A web client to build workflows and manage the processing
is currently being built.
BFIO
32
is a Python library that supports reading of all
160 + Bio-Formats supported file formats, with opinionated
but highly optimized reading and writing of OME-TIFF and
OME-Zarr for use in large scale applications. All changes
and updates to the specification are implemented in the
library. Similar to AICSImageIO, BFIO can act as a format
conversion tool where it is possible to load data in diverse
image formats and immediately pass the NumPy array to
a custom OME-Zarr writer function. BFIO distinguishes
itself from other NGFF loaders in that it has focused on
performance and performs chunked data read/writes to load
and save images in an out-of-core fashion by default. BFIO
is opinionated about chunk size and loading/saving pattern
and this loss of freedom by the user allows BFIO to make
substantial gains in its read/write speeds. BFIO is currently
in the process of being refactored to utilize TensorStore with
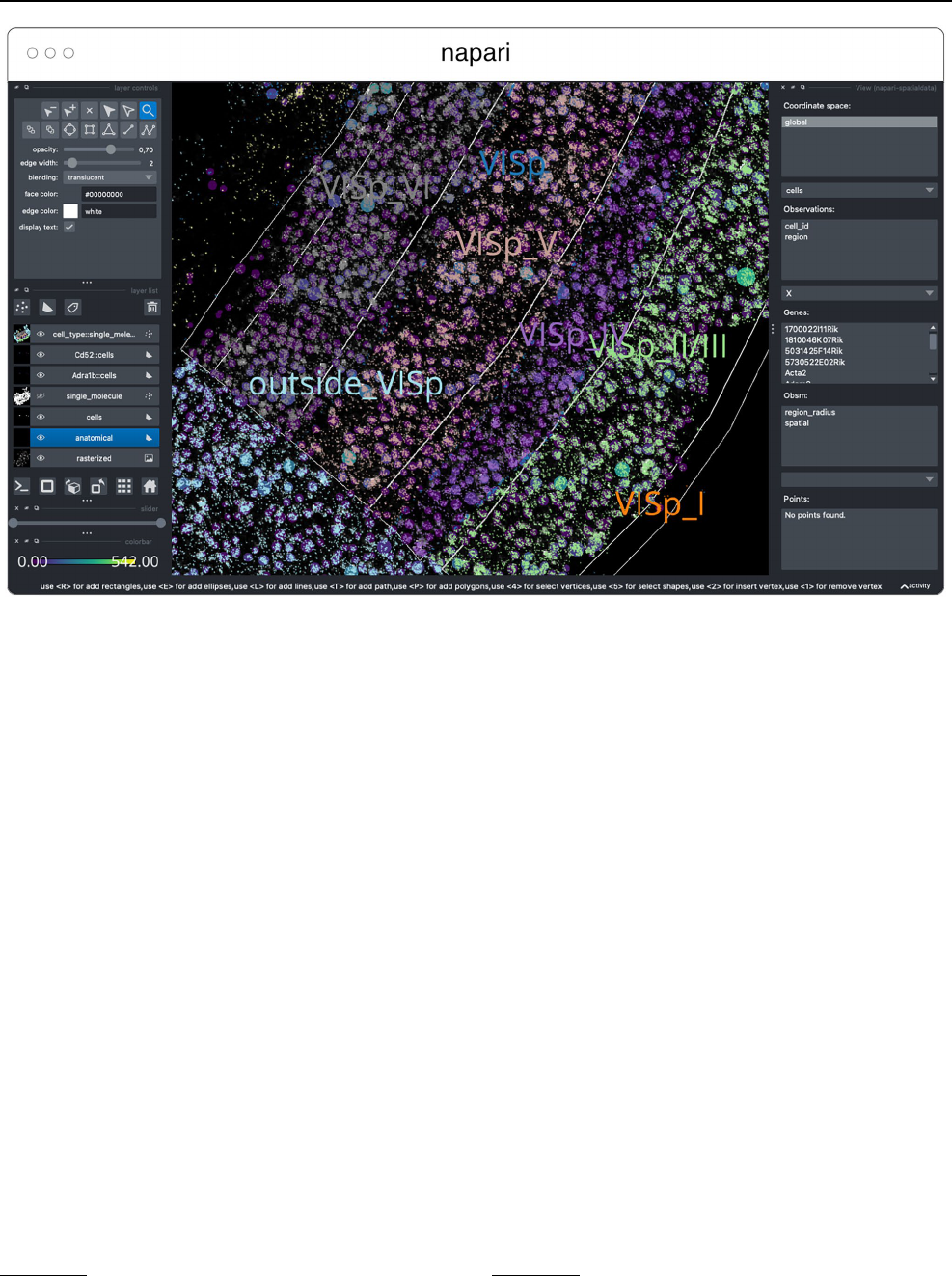
Fig. 10 Example application of BigStitcher’s interest point based reg-
istration on one of the first “exaSPIM” lightsheet microscope datasets
acquired at the Allen Institute for Neural Dynamics (sample 609,281,
available at the link in Table2). The overlapping regions of two tiles
are shown in BDV at their nominal (left) and aligned (right) loca-
tions. This large scale NGFF dataset consists of 54 tiles with dimen-
sions of 24,576 × 10,656 × 2048 voxels (about 1TB raw size) each
28
https:// ome- zarr. readt hedocs. io/.
29
https:// fract al- analy tics- platf orm. github. io.
30
https:// slurm. sched md. com/ docum entat ion. html.
31
https:// www. youtu be. com/ watch?v= DfhRF 1OW5CE.
32
https:// github. com/ Polus AI/ bfio.
Histochemistry and Cell Biology
1 3
subsequent additions to TensorStore to support the full OME
data model, and is planned to release the optimized Tensor-
Store read/write in the coming months.
SpatialData
33
is a Python library that provides an on-disk
format and an in-memory object for working with spatial
omics data (Fig.11). It uses the ome-zarr-py readers and
writers for raster data (images and labels) and implements
features of the upcoming version of the NGFF specifica-
tion, including tables and transformations. Furthermore,
it experimentally supports the representation of additional
modalities commonly found in spatial omics data, such as
points (e.g. transcripts locations) and shapes (e.g. spatial
transcriptomics circular array capture locations and generic
polygonal ROIs). The library implements a set of operations
such as aggregation of molecular measurements (associating
transcripts locations to cells) and efficient spatial queries that
are interoperable across representations. Finally the Spatial-
Data framework provides readers for datasets produced by
the most popular spatial omics technologies, a static plot-
ting library based on matplotlib (Hunter 2007) as well as a
napari plugin for interactive visualization and annotation of
the data.
C++
TensorStore
34
is an open-source C++ library and Python
wrapper that provides a unified, high-performance interface
for accessing a variety of array formats, including Zarr. In
addition to use with scientific imaging data, it is also used
to store and load checkpoints of massive machine learning
models with hundreds of billions of parameters. Supported
underlying storage mechanisms include: arbitrary HTTP
servers (read-only); local and network filesystems; and
Google Cloud Storage. It supports safe, concurrent write
access from multiple machines, without the need for a sepa-
rate lock server, through the use of optimistic concurrency.
While it does not yet specifically support OME-Zarr multi-
scale metadata, it can be used to read and write individual
scale levels as Zarr arrays. Specific abstractions for multi-
resolution data, along with support for OME-Zarr multiscale
metadata, is planned.
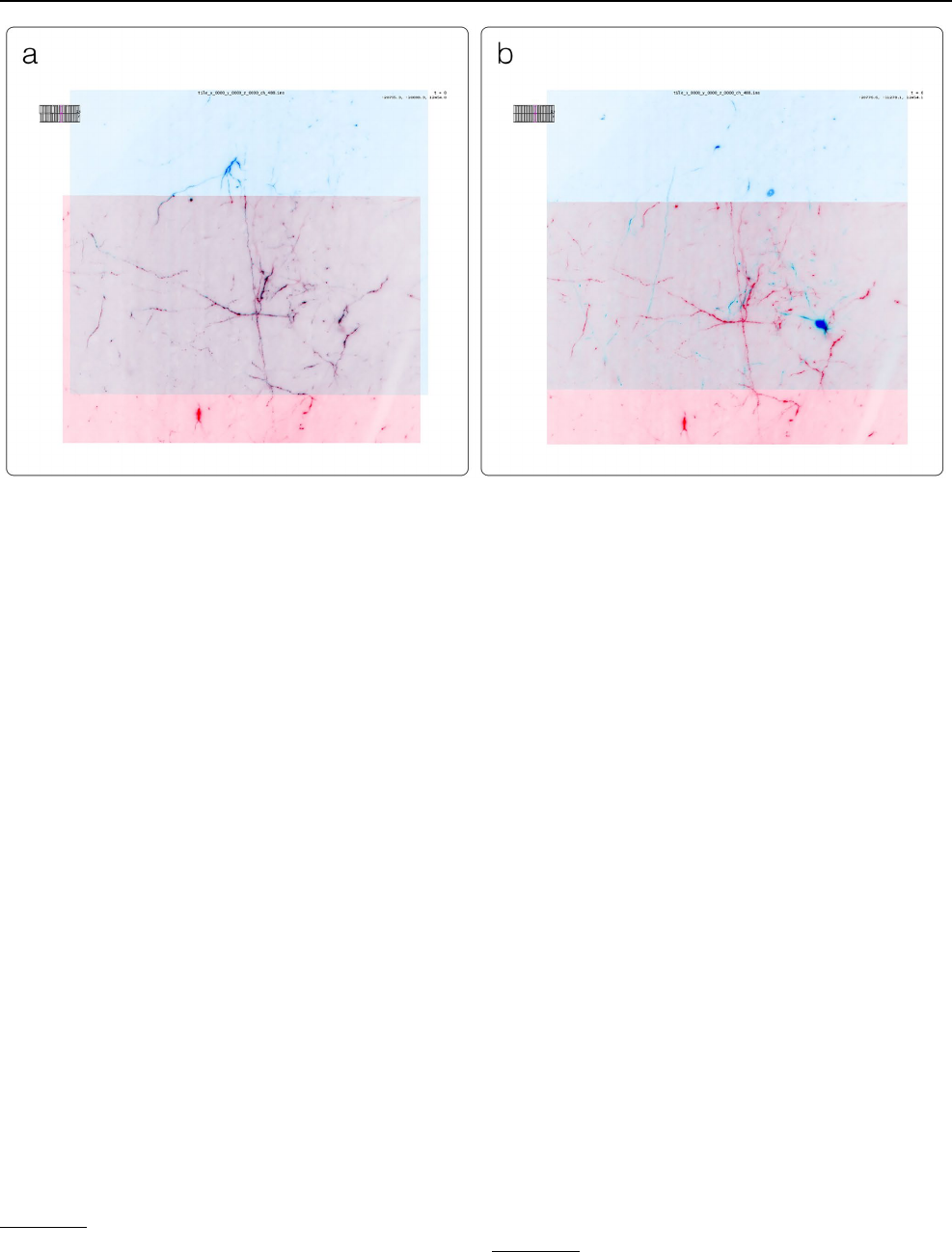
Fig. 11 Visualization of a MERFISH mouse brain dataset [Allen
Institute prototype MERFISH pipeline (Long et al. 2023)] via the
napari-spatialdata plugin, featuring single-molecule transcripts
(points) and their rasterized representation (image), polygonal ROIs,
and annotated cells approximated as circles with variable radii. The
dataset has been converted to OME-Zarr with the SpatialData APIs
33
https:// spati aldata. scver se. org/ en/ latest/.
34
https:// google. github. io/ tenso rstore/.

Histochemistry and Cell Biology
1 3
Nyxus
35
is an open-source C++, plus Python wrapper
available on Conda or PyPI, that provides a high perfor-
mance and out-of-core, parallel feature extraction library
for image and annotation data natively in OME-Zarr. The
library assumes that regions of interest are labeled and can
be stored as an annotation layer within OME-Zarr files or
stored as separate OME-Zarr files from the raw intensity
data. Nyxus supports feature extraction on 2-dimensional
or 3-dimensional data and contains more morphological,
histogram, and texture features than most individual librar-
ies. It can compute these features in an out-of-core fashion
enabling users to analyze images/volumes of unlimited size.
Nyxus supports whole image or typical region based feature
extraction, it also supports nested feature extraction (i.e. par-
ent annotation with children annotations) and a single region
across many channels.
Java
For reading OME-Zarr data with Bio-Formats in Java, the
OMEZarrReader
36
has been developed. Once installed, this
plugin allows opening OME-Zarr images from any applica-
tion that currently makes use of Bio-Formats plane-based
API. This includes ImageJ or RBioFormats for accessing
OME-Zarr data in R.
The N-dimensional N5 API supports reading array data
and arbitrary metadata using Zarr, HDF5 and the N5 for-
mat. Its associated Fiji plugins
37,38
, can therefore read pixel
/ voxel data from OME-Zarr containers, but not its metadata
specification at the time of this writing. The developers have
committed to adding OME-Zarr support in the future by
developing a shared implementation with MoBIE. Support
for multiple backends makes the N5 API an appealing choice
for extending NGFF support to other, write-optimized stor-
age formats.
Example
An example analysis of a dataset from the Image Data
Resource (IDR) (Williams etal. 2017) helps to illustrate
the possibilities available to the end-user from an analysis
perspective. A light sheet fluorescence microscopy image
published by McDole etal. (idr0044)
39
is composed of 2
channels, approximately 1000 z-sections and 500 timepoints.
Each 2D-plane is of dimension 2k × 2k. Numerous z-sec-
tions were acquired but the relevant planes are the middle
z-sections. Such data is particularly useful for teaching and
training purposes, so it is usually only necessary to access a
limited subset of an image.
Traditionally, two options are available in order to analyze
an image. One can download the full image. This approach
is far from ideal since only a portion of the data is required
for analysis. Additionally, a specific image reader is needed
to interpret the data, potentially limiting which analysis
language/framework could be used to perform the analysis.
Alternatively, the relevant planes could be retrieved using
the IDR Application Programming Interface (API). The IDR
API is very versatile but it complicates the parallelisation
of tasks for users. To enable streamable access by all of the
tools and libraries outlined above, the images of the study
were converted using bioformats2raw (see the “Generators”
section) and made available on object storage at EBI (See
Table3).
The nature of OME-Zarr allows the end-user to take full
advantage of libraries like Dask
40
, a free and open-source
parallel computing library that scales the existing Python
ecosystem. An analysis task like segmentation is broken into
many individual steps that are run independently. Each step
lazily loads the chunk of the image that it is to work on,
and then the result is aggregated. Available public resources
like Google Colab
41
or mybinder
42
and publicly accessible
Table 3 Comparison of access methods to data stored in the IDR
OME-Zarr provides the fastest and most flexible access when accessing less than an entire dataset. A notebook documenting the OME-Zarr
access is available at https:// github. com/ ome/ omero- guide- python/ blob/ master/ noteb ooks/ idr00 44_ zarr_ segme ntati on_ paral lel. ipynb
Download (via Aspera) IDR API access (via Ice) OME-Zarr access (via S3)
Load image subregion, e.g.,
single chunk or tile
No, only per file Yes Yes
Lazy loading No No Yes. Use Dask collections:
da.from_zarr (endpoint_url)
Easily analyze in parallel No, depends on file format which may
require a translation library
Difficult due to the transfer proto-
col used (zeroc-ice)
Yes. Use Dask schedulers:
dask.delayed (analyze) (t, c, z)
35
https:// github. com/ Polus AI/ nyxus.
36
https:// github. com/ ome/ ZarrR eader.
37
https:// github. com/ saalf eldlab/ n5- ij.
38
https:// github. com/ saalf eldlab/ n5- viewer.
39
https:// idr. openm icros copy. org/ webcl ient/? show= proje ct- 502.
40
https:// www. dask. org/
41
https:// colab. resea rch. google. com/.
42
https:// mybin der. org/.

Histochemistry and Cell Biology
1 3
data were sufficient for training purposes but the approach
could easily be extended for larger scale analysis. This facili-
tates training the next generation of scientists on how to use
cloud-computing resources.
Generators
For the foreseeable future, datasets will exist in one of the
many forms they exist in today. As outlined in (Moore etal.
2021), translating those on the fly brings delays in visualiza-
tion and analysis that can be solved by performing a single
conversion to OME-Zarr. This process captures all metadata
that Bio-Formats is aware of in an open format, and decou-
ples the user from the version of the vendor software used to
capture the data. Several generation tools (Table4) are avail-
able based on the particular environment you are working in.
Bioformats2raw
Bioformats2raw
43
is the original command-line utility to
convert various image file formats into OME-Zarr format.
Bioformats2raw offers rich and flexible parameter options,
giving the user extensive control over the conversion process
as well as freedom to specify various features of the output
Zarr datasets. A few of the interesting input parameters are
the chunk dimensions, the number of resolution levels, the
compression algorithm, and the number of workers. Bio-
formats2raw can read all proprietary file formats supported
by Bio-Formats as well as a select few file format readers
supported only in bioformats2raw, including the 3D-Histech.
mrxs format. The input, therefore, can be single or multiple
series as well as high-content screening (HCS) data. The
conversion will be performed according to the respective
OME-NGFF specification. Multiscaling is achieved either
by creating sub-resolutions during the conversion process,
or by using the existing ones from the input format.
Bioformats2raw is optimal for remote and headless opera-
tion and can be conveniently built into pipelines, e.g., by
using workflow management systems, such as Galaxy, Nex-
tflow, Snakemake, etc. that would also facilitate parallel
conversion of batches of image data into OME-Zarr, for
example on HPC clusters. Additionally, cloud-aware tools
like Distributed-OMEZarrCreator (Weisbart and Cimini
2022) allow easy wrapping of bioformats2raw on Amazon
Web Services (AWS). By default, bioformats2raw writes an
OME-XML metadata to a specific directory in the output
Zarr hierarchy. This metadata can then be used by a comple-
mentary package, namely raw2ometiff, to convert the output
from bioformats2raw into OME-TIFF.
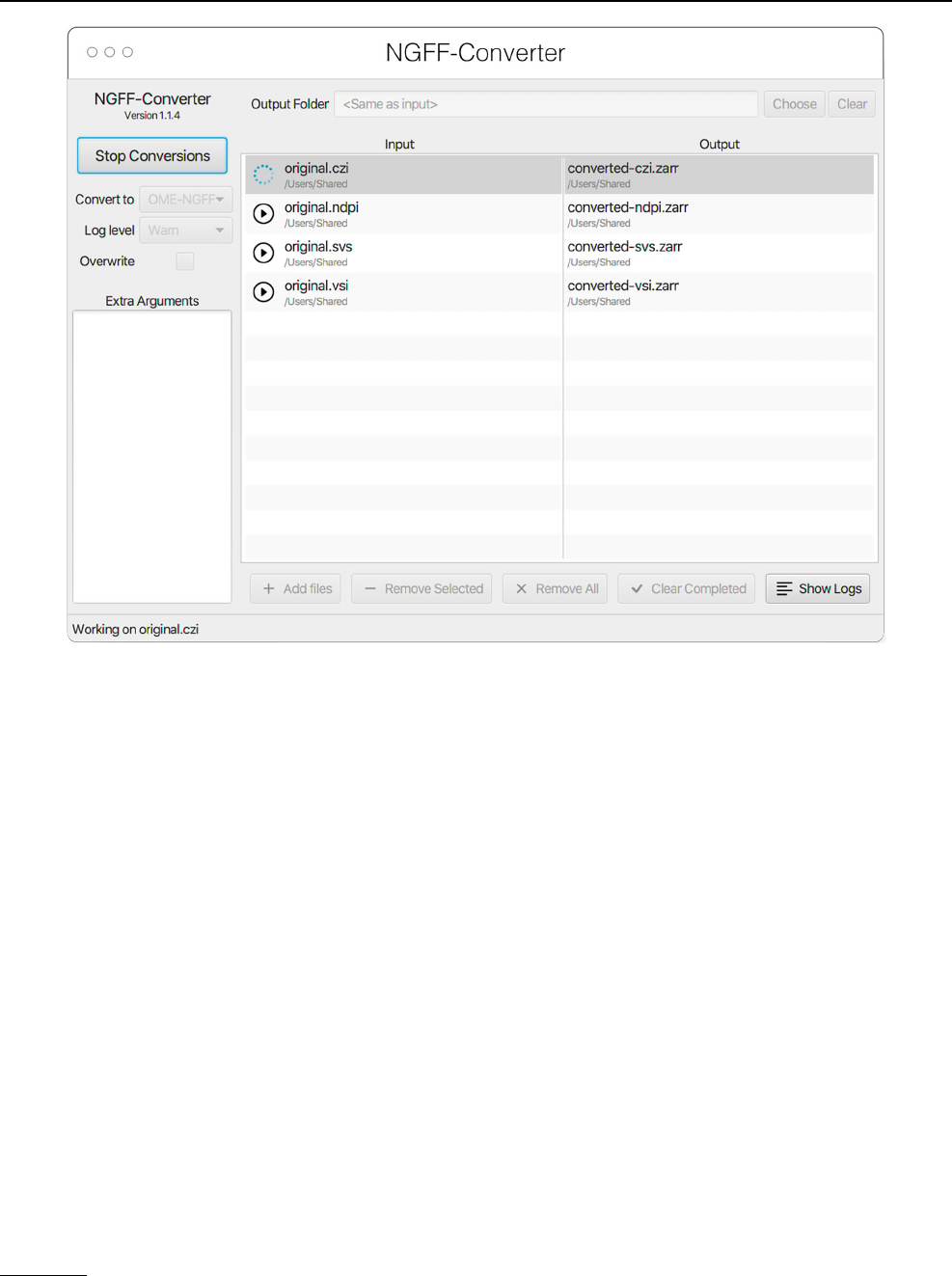
NGFF‑Converter
Glencoe Software’s NGFF-Converter
44
(Fig.12) is an
easy-to-use and intuitive graphical user interface (GUI)
application supporting conversion of any format readable
by Bio-Formats, as well as the additional readers built into
bioformats2raw. By packaging the command line utili-
ties bioformats2raw and raw2ometiff, NGFF-Converter
can convert numerous file formats to both OME-TIFF and
OME-Zarr based on the user selection. NGFF-Converter is
approachable for users less familiar with command line utili-
ties while maintaining the flexibility of tunable parameters
as described in the previous section. In addition, NGFF-
Converter was developed with batch processing in mind,
supporting the scheduling of multiple conversions with clear
visuals of conversion job status. NGFF-Converter is avail-
able for both Windows and MacOS.
ImSwitch
At the same time, we envision an increasing number of hard-
ware devices capable of directly outputting image data in
OME-Zarr, streamlining analysis and reducing the risk of
data duplication. One example of software that facilitates
this approach is ImSwitch
45
(Casas Moreno etal. 2021), a
modular open-source microscope control software written
in Python. Imswitch implements an architecture based on
the model-view-presenter design pattern to enable flexible
and modular control of multiple microscope modalities.
Table 4 List of software for
generating OME-Zarr data in
the order they are described
below along with a brief
description of their use
An up-to-date version of the table is maintained at https:// ngff. openm icros copy. org/ tools and contributions
are welcome
Generator Use Comment
bioformats2raw Command-line, Java Scriptable tool for integration into workflows
NGFF-Converter Windows, MacOS Graphical user-interface with a queue of files
ImSwitch Python Stream data from hardware
Kerchunk Python Preprocess non-Zarr files to simulate OME-Zarr
43
https:// github. com/ glenc oesof tware/ biofo rmats 2raw.
44
https:// www. glenc oesof tware. com/ produ cts/ ngff- conve rter/.
45
https:// imswi tch. readt hedocs. io/ en/ stable/.
Histochemistry and Cell Biology
1 3
The experimental data acquired, with related experiment
metadata, can be directly written with ome-zarr-py into
OME-Zarr files. The file format showed to be favorable
when using workflows involving distributed computational
resources for processing (e.g. parallel RESOLFT reconstruc-
tions) of image data (Casas Moreno etal. 2023). Previously
used acquisition parameters and settings can be loaded into
ImSwitch from saved data to conveniently enable reproduc-
ible workflows.
Kerchunk
Within the Python ecosystem, it is also possible to “simu-
late” an OME-Zarr dataset with kerchunk
46
. This library
pre-processes existing data formats like TIFF and HDF5
to generate a JSON file containing all metadata and chunk
locations. Support for other file formats can be added if
each chunk can be represented by a combination of path to
a file, location in that file, and length of the chunk. Using
this mechanism, it is possible to leave data in a monolithic
format but still achieve some of the benefits of OME-Zarr.
Support in other programming languages is possible based
on community interest.
Other ways tocreate OME‑Zarr
In addition to using these dedicated generators, many of the
general-purpose tools mentioned also support the generation
of OME-Zarr data. Within the Fiji ecosystem the MoBIE
plugin provides a GUI for creating OME-Zarr. Among its
other functionalities, MoBIE can convert images imported
by Fiji into OME-Zarr. The input is imported via Fiji read-
ers, which include Bio-Formats, and enables immediate
visualization and exploration options of the OME-Zarr data.
All uploads to webKnossos from all supported formats are
also automatically converted into OME-Zarr, which can be
streamed or downloaded for use with other tools. Libraries
like ome-zarr-py can write numpy and Dask arrays to OME-
Zarr according to the OME-NGFF specification. Where
users are already manually handling the reading of the input
data and the parsing of the metadata in Python code, this
may be the easiest path to generating OME-Zarr data.
Fig. 12 NGFF-Converter GUI showing a sample of input formats being converted to OME-Zarr
46
https:// github. com/ fsspec/ kerch unk

Histochemistry and Cell Biology
1 3
Examples ofshared OME‑Zarr data
Not just individual users or research projects are faced with
the issues of format compatibility. Large-scale bioimag-
ing resources are also moving to OME-Zarr to ease access
across a range of storage options. Below we discuss several
of the ways in which these and other institutions are sharing
their data with the OME-Zarr format as examples of what
is possible as well as where you might find existing data
today, all summarized in Table5. However, as with the tools
above, these and other resources are being actively updated.
Users interested in re-using datasets can refer to https:// ngff.
openm icros copy. org/ data for an up-to-date version of this
table maintained by the community or submit new resources
as they become available. Though central registry does not
exist for other file formats, the ease of access to OME-Zarr
on the web, e.g. through embedded multiple-terabyte data in
a static webpage, makes such a catalog particularly valuable
and the growing availability of OME-Zarr formatted data
will hopefully accelerate tool development.
Amazon S3
Some of the most visible uses of OME-Zarr are part of the
“Public Data” programs provided by large, commercial
vendors like Amazon
47
and Google
48
to share commu-
nity-critical datasets. Submissions to these programs are
reviewed for overall value to the community, but if accepted,
represent a particularly accessible resource.
Cell painting gallery
The Cell Painting Gallery
49
contains Cell Painting (Bray
etal. 2016) images and image-based profiles from many
publicly available datasets, hosted by AWS Open Data Reg-
istry. Currently, the LINCS Dataset (Way etal. 2022) is
available in the Cell Painting Gallery in OME-Zarr format.
In LINCS, 110,012,425 A549 human lung carcinoma cells
across 136 plates were treated with 1,571 compounds across
6 dose points. Morphology was captured by a standard Cell
Painting workflow of five fluorescent channels covering
eight organelles. Image data was converted to OME-Zarr
using bioformats2raw with the Distributed-OMEZarrCreator
wrapper (Weisbart and Cimini 2022). 1,790 morphological
measurements were taken using CellProfiler (Kamentsky
etal. 2011) which are also available in the Cell Painting
Table 5 List OME-Zarr resources data belonging to the authors, broken down by storage types in the order they are described below along with
rough estimates of their size at the time of publication
These include catalogs and dashboards which will help the reader discover datasets as well as some resources which are migrating to OME-Zarr.
An up-to-date version of the table is maintained at https:// ngff. openm icros copy. org/ data and contributions are welcome
Storage Catalog Dashboards/datasets Zarr files Size
Amazon S3 Cell painting gallery https:// github. com/ broad insti tute/ cellp ainti ng- galle ry 136 20TB
DANDI https:// dandi archi ve. org/ dandi set/ 000108
https:// ident ifiers. org/ DANDI: 000108
https:// github. com/ dandi sets/ 000108
3914 355TB
Neural dynamics https:// regis try. opend ata. aws/ allen- nd- open- data/ 90 200TB
Glencoe https:// glenc oesof tware. com/ ngff 8 165GB
Alternative S3
providers
BIA samples https:// bit. ly/ bia- ome- ngff- sampl es 90 200GB
IDR samples https:// idr. github. io/ ome- ngff- sampl es/ 88 3TB
Sanger https:// www. sanger. ac. uk/ proje ct/ ome- zarr/ 10 1TB
SSBD https:// ssbd. riken. jp/ ssbd- ome- ngff- sampl es 12 196GB
On-premise CZB-Zebrahub https:// zebra hub. org 5 1.2TB compressed
MoBIE https:// mobie. github. io/ specs/ ngff. html 21 2TB
SpatialData https:// github. com/ scver se/ spati aldata- noteb ooks/ tree/
main/ datas ets
10 25GB
webKnossos https:// zarr. webkn ossos. org 69 70TB
In-progress
Brain Image Library https:// www. brain image libra ry. org/
HuBMAP https:// portal. hubma pcons ortium. org/
OpenOrganelle https:// openo rgane lle. janel ia. org/ datas ets
47
https:// aws. amazon. com/ opend ata/ open- data- spons orship- progr
am/.
48
https:// cloud. google. com/ stora ge/ docs/ public- datas ets.
49
https:// regis try. opend ata. aws/ cellp ainti ng- galle ry.

Histochemistry and Cell Biology
1 3
Gallery. More Cell Painting datasets in the Cell Painting
Gallery are planned for both conversion to OME-Zarr and
browsability through IDR.
DANDI
DANDI, “Distributed Archives for Neurophysiology Data
Integration” is supported by the US BRAIN Initiative as an
open-access data repository for publishing and sharing neu-
rophysiology data including electrophysiology, optophysi-
ology, and behavioral time-series, as well as images from
immunostaining experiments. As datasets get larger, down-
loading whole datasets for analysis or visualization becomes
increasingly impractical. Thus, DANDI allows data to be
operated on in the cloud using standardized methods and
computational servers near the data. It enforces the organ-
ized structure of the Brain Imaging Data Structure (BIDS)
and its microscopy extensions (Bourget etal. 2022), with
the latest specification
50
allowing such data to be stored in
the OME-Zarr format. DANDI uses standardized metadata
(JSON, JSON-LD), data organization (BIDS, or BIDS-like
subset), and data storage formats (e.g., OME-Zarr) that
allow the metadata to be queried before data access, and
data to be accessed partially and at different resolutions.
The archive stores open and embargoed data on Amazon
S3 in the US and is supported by the Amazon Open Data
program. The DANDI API has full programmatic support
for uploading and accessing Zarr objects, and researchers
can use the DANDI command line interface tool to upload
BIDS organized Zarr data to DANDI. All datasets are also
available as DataLad (Halchenko etal. 2021) datasets from
GitHub
51
with individual Zarr objects provided as separate
repositories
52
and linked to original datasets as git submod-
ules. Since data itself resides on the versioned S3 bucket,
such DataLad datasets provide access to TBs of OME-Zarr
data with unambiguous versioning.
DANDI allows any researcher to view or compute on
the data. Lightly pre-processed data and metadata are
being made available via DANDI for the BRAIN Initia-
tive and other projects. Two such datasets were produced
by the BRAIN Initiative Cell Census Network (biccn.org)
comprising 370TB of data. About 60% of a single human
brain hemisphere is available in a dataset with vascular,
neuronal, and cellular stains (290TB; an example 3D slab
shown in Fig.13) with an additional 65TB from the brains
of two other participants. These data can be visualized by
anyone on the planet due to horizontal scalability provided
by AWS S3, the use of a streaming data storage format
(OME-Zarr), and interactive visualization tools such as
Neuroglancer, ITK-VTK-Viewer, and others described ear-
lier. For the OME-Zarr data, each such object provides an
option to directly open the object in the ITK-VTK-Viewer
or for checking using the OME-Zarr Validator. To optimize
data transfer, DANDI recommends OME-Zarr lightsheet
imaging data on human brains to be stored with chunk
sizes of (1,1,128,128,128) and with lossless Blosc + Zstd
compression.
Others
A number of other projects like Allen Institute for Neural
Dynamics (AIND)
53
have also followed this strategy to
achieve FAIR, Open and Reproducible Science. However,
it is equally possible to host your own data on commer-
cial services if funds are available. Glencoe Software, as
the commercial arm of OME, regularly stores OME-Zarr
data in buckets like s3://gs-public-zarr-archive
54
. A careful
consideration is needed of the price per terabyte as well as
egress costs incurred by users accessing such buckets.
Alternative S3 providers
S3 has become a de facto standard API. As such, a num-
ber of providers offer commercial systems with many of the
same features. Several publicly funded institutions dedicate
themselves to providing large-scale infrastructure for shar-
ing biological data and this is often achieved by providing
“S3-like” storage.
BIA andIDR
An example of an institute providing a large-scale cloud
repository is the European Bioinformatics Institute (EBI)
who has done so by investing in object, or “cloud”, storage.
The Image Data Resource (IDR) (Williams etal. 2017) and
the BioImage Archive (BIA) (Hartley etal. 2021) are free
public digital repositories for biological images hosted at
EBI that are associated with peer-reviewed publications or
are of value beyond a single experiment. The BIA strives
to provide hosting for all such publications while the IDR
focuses on a selection of reference datasets which are further
curated. They accept submissions of many different image
formats to enable easy and fast sharing. However, most of
these formats have issues relating to metadata and to online
visualization as discussed above.
50
https:// bids- speci f icat ion. readt hedocs. io/ en/ stable/ 04- modal ity-
speci fic- files/ 10- micro scopy. html# file- forma ts.
51
https:// github. com/ dandi sets.
52
https:// github. com/ dandi zarrs.
53
https:// regis try. opend ata. aws/ allen- nd- open- data/.
54
https:// glenc oesof tware. com/ ngff.
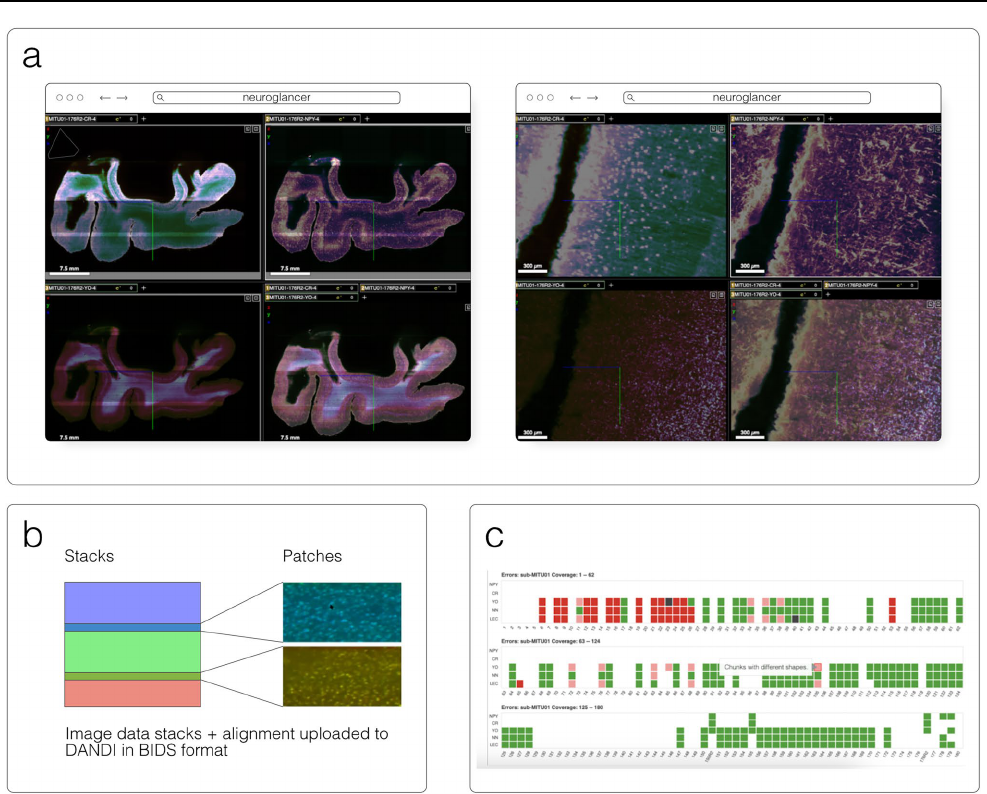
Histochemistry and Cell Biology
1 3
The IDR’s need to store data on object storage led OME
to begin discussions around a potential NGFF. As early
adopters and implementers, the IDR found that such a file
format alleviated many bioimaging bottlenecks at scale.
Multi-terabyte 3D volumes (McDole etal. 2018) and 100
terabyte high-content cell painting screens (Bray etal. 2016),
common to the IDR, can be more reasonably processed and
accessed if converted in the repository to OME-Zarr. Addi-
tionally, use of OME-Zarr as a submission format reduced
the burden on the curation team to deal with compatibility
of custom formats.
In the BIA, OME-Zarr is chosen as its chunked architec-
ture and available libraries and visualization tools enable on-
the-fly visualization of the data, including 3D images, time
series data and images with multiple channels. Moreover, it
can accommodate extensive image level metadata, and has
substantial community support.
To enable easier sharing and investigation of long-term
solutions, images are converted from selected datasets,
including most machine learning datasets, into OME-Zarr
format. The converted representation is stored transiently
on fast, S3 compatible storage. This enables serving the
images for on-the-fly visualization in embedded viewers,
visualization by desktop clients that can access OME-Zarr
remotely such as napari or MoBIE, and provides fast access
for on-the-fly analysis via Jupyter notebooks. The converted
image representations are transient due to the need to bal-
ance resources between cheaper scalable storage for long
Fig. 13 An OME-Zarr dataset on DANDI (ID: DANDI:000108) con-
tains multiscale 5D datasets (time, channel, z, y, x), with metadata,
scale, and transformations. a DANDI:000108 includes multi-slab,
multi-stain (NeuropeptideY-NPY, Calretinin-CR, YOYO1) data that
are aligned and the coordinate transforms are stored in the Zarr files,
allowing b on-the-fly visualization and stitching of the slabs at multi-
ple scales (two shown) using Neuroglancer. c An HTML dashboard
allows data submitter and any user on the Web to see the live status:
samples + stains uploaded, their quality issues, and multiscale view-
ing of this 300 + TB dataset

Histochemistry and Cell Biology
1 3
term archival and fast storage for access to in-demand data-
sets. The OME-Zarr converted image representations are
currently presented on separate websites, with plans to inte-
grate these within the primary dataset pages. Images can be
displayed using Vizarr, and an option to view them with the
ITK viewer is also provided. As with all submissions, each
image representation has a unique URI therefore the users
can access and view them with their favorite online view-
ers, without the need to download. Already it is possible to
submit datasets that have been converted into OME-Zarr and
as this solution matures, submitters will be encouraged to
pre-convert complicated datasets to lower the overhead on
the curation teams.
Sanger
Data generated by Wellcome Sanger Institute and Newcastle
University are made available on their public S3 buckets.
That includes both array-based single cell sequencing data
and microscope-based imaging data. The data conversion
was performed via a bespoke data ingestion pipeline which
translates every object into OME-Zarr, chosen to be the
default image format for all the data generated from spatial
technologies such as Visium, insitu sequencing, RNAscope,
Xenium and Merfish.
SSBD
SSBD is a platform for Systems Science of Biologi-
cal Dynamics to share and reuse bioimaging data hosted
at RIKEN, Japan (Tohsato etal. 2016). It consists of two
systems; SSBD:database for sharing highly reusable data
with rich metadata and SSBD:repository for rapid sharing
of all bioimaging data in journal papers. To support the
development and validation of software tools for NGFF, we
converted selected datasets in SSBD:database into OME-
Zarr format using the bioformats2raw converter. The data-
sets include those obtained with light sheet fluorescence
microscopy, STED super-resolution microscopy, FIB-SEM,
and other state-of-the-art microscopies. They are currently
stored and available on S3-compatible storage in the RIKEN
HOKUSAI SS system.
On‑premise
Even beyond conventional object storage, OME-Zarr pro-
vides a mechanism for consistently sharing bioimaging data.
This can be as simple as putting the files on an existing web
server or dedicated servers can be installed locally.
Zebrahub
Zebrahub
55
is a comprehensive atlas of zebrafish devel-
opment (Lange etal. 2023) paired with a fully interactive
website to explore both single-embryo single-cell RNA
sequencing datasets and light-sheet microscopy time-lapses.
Light-sheet microscopy is a powerful imaging technique
that, when combined with transgenic zebrafish expressing
fluorescent proteins, can reconstruct the entire embryonic
development at a cellular level. This results in terabyte-scale
time-lapse datasets that contain 3D images acquired at regu-
lar intervals, often every 30s. Acquiring multiple channels
is necessary to answer specific biological questions, which
further increases the data's size and dimensionality. Stor-
ing, processing, visualizing, and disseminating such data
becomes challenging. The OME-Zarr format addresses
several of these issues by providing a well-established and
maturing standard storage format supported by various
image processing and visualization tools. The Zebrahub
project and its accompanying website leverage OME-Zarr
by providing image data downloads in OME-Zarr format and
neuroglancer instances that allow for the interactive explora-
tion of these image datasets (see Fig.14). Sharing terabyte-
scale high-resolution light-sheet imaging time-lapse datasets
in a web browser via OME-Zarr is a significant contribution
towards advancing observation-based discovery in develop-
mental biology.
Hosting your own
Alternatively there are existing software solutions that expose
an object-storage interface to files on disk like Minio
56
. The
European Molecular Biology Laboratory in Heidelberg is
currently hosting all of the reference data for the develop-
ment of the MoBIE tool by running Minio on their own
hardware. Examples of datasets formatted in OME-Zarr
include https:// git hub. com/ mobie/ covid- if- proje ct, https://
github. com/ mobie/ htm- test and https:// github. com/ mobie/
spati al- trans cript omics- examp le- proje ct. In the README
of those repositories, a list of all S3 OME-Zarr URLs can be
found which can be opened with viewers other than MoBIE.
Sample data for the SpatialData library can also be found on
EMBL’s S3 resource. A list is available under https:// github.
com/ scver se/ spati aldata- noteb ooks/ tree/ main/ datas ets.
Alternatively, dedicated, bioimaging server solutions are
available for hosting and serving OME-Zarr data. As men-
tioned above, webKnossos makes all datasets accessible in
the OME-Zarr format. To import data into webKnossos a
conversion is required, whether automatically on browser
55
http:// zebra hub. org/.
56
https:// min. io
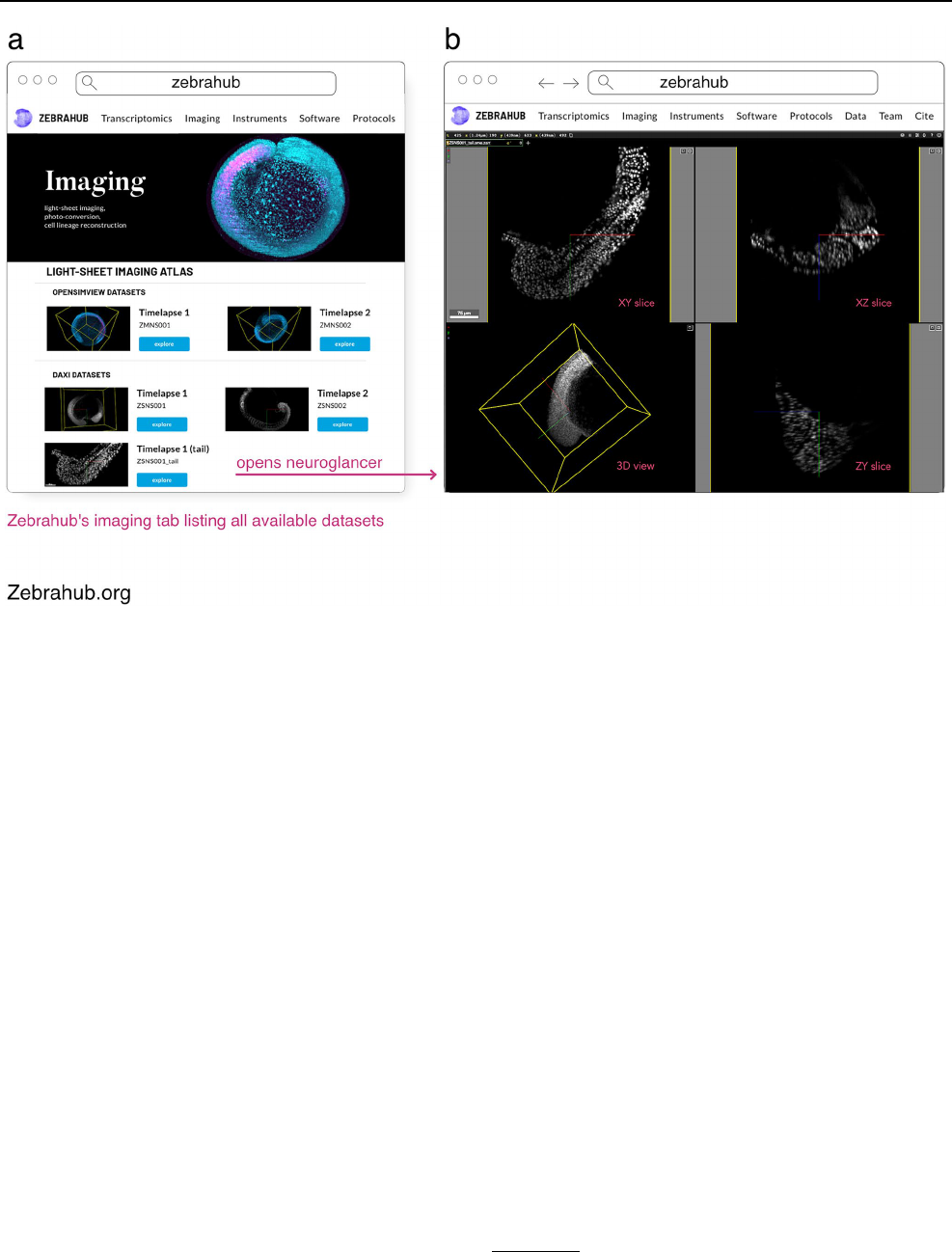
Histochemistry and Cell Biology
1 3
upload or manual through filesystem import to OME-Zarr or
WKW, the “webKnossos Wrapper” format. Once imported
into webKnossos, datasets can be made publicly available
via the dataset settings. This is an easy way to publish data-
sets directly from an institutional HPC storage. A collec-
tion of published OME-Zarr data is available at https:// zarr.
webkn ossos. org/.
Another possible mechanism for hosting OME-Zarr data
is OMERO (Allan etal. 2012). The OMEZarrReader plugin
to Bio-Formats is included in OMERO since version 5.6.5,
enabling import of OME-Zarr data into any OMERO includ-
ing the IDR. These are stored in OMERO’s internal reposi-
tory with other file types and appear as images, or in the case
of HCS data plates, in the standard hierarchy. Users can then
use the OMERO API or clients (OMERO.figure, QuPath, or
even R) to access the data. To enable OME-Zarr access to
end users, services are available which expose an OME-Zarr
compatible endpoint to clients.
In‑progress
Finally, several repositories have plans of migrating all of
their existing datasets to OME-Zarr as a common represen-
tation for dissemination.
Brain Image Library
Institutes with large scale parallel (HPC) file systems like
the Pittsburgh Supercomputing Center, home to the Brain
Image Library
57
(BIL), are building tools to share OME-Zarr
formatted data without object storage and without modify-
ing the existing data repository
58
. Still in early development,
these services will make 1000s of whole brain microscopy
datasets, including murine, NHP and human, some expected
to be over a petabyte in size, from the BICCN (BRAIN
Initiative Cell Census Network) (BICCN Data Ecosystem
Collaboration etal. 2022) and BICAN (BRAIN Initiative
Cell Atlas Network) available in OME-Zarr where appro-
priate starting this year. To achieve this, microservices will
automatically provide an OME-Zarr representation of the
day on the fly to prevent data duplication. These microser-
vices will enable all of the visualization software described
above (Fig.15) to make use of the data while permitting the
high-performance computing facilities to optimize for their
Fig. 14 a ‘Imaging’ tab of the zebrahub.org website showing the list of imaging datasets made available. b By clicking on a particular dataset,
the user is directed to a neuroglancer instance that allows interactive exploration of the dataset
57
https:// www. brain image libra ry. org/.
58
https:// github. com/ CBI- PITT/ BrAin PI
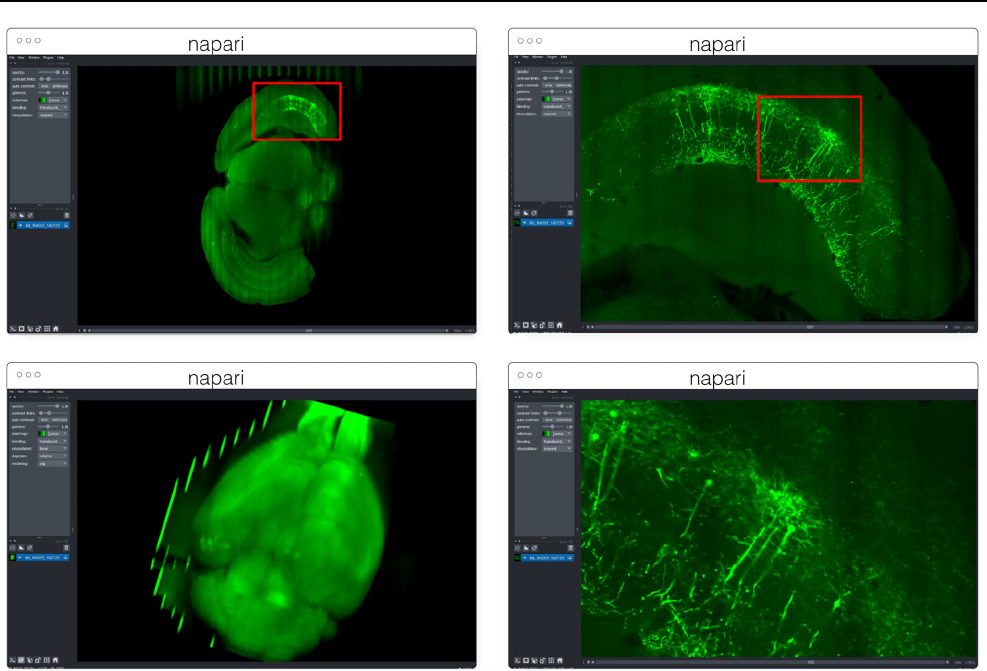
Histochemistry and Cell Biology
1 3
storage requirements. Similar APIs will be employed locally
at BIL to allow HPC computational tools to interface with
the data in a standard way, and with community derived
tools developed around OME-Zarr.
HuBMAP
The HuBMAP (“Human Biomolecular Atlas Program”)
consortium (HuBMAP Consortium 2019) adopted open-
standard imaging formats and developed novel web-based
technologies in order to simplify the server-side infrastruc-
ture traditionally required to support visualization of such a
bioimaging resource. Consideration of infrastructure costs
and a commitment to FAIR data access principles led the
consortium to adopt OME-TIFF as an internal standard.
In conjunction, members of the consortium developed Viv
(Manz etal. 2022)—a client-side bioimaging visualization
library—to remove a dependency on server-side render-
ing and enable flexible browsing of multi-terabyte datasets
directly within the HuBMAP data portal website. Currently,
HuBMAP is working to adopt OME-Zarr in order to support
a wider diversity of modalities (e.g., large 3D volumes) as
well as integrate additional data types (e.g., segmentations,
ROIs, 3D rendering, high-content screening data, and non-
image formats like AnnData) not supported by OME-TIFF.
This data will be hosted on the Pittsburgh Supercomputing
Center’s hardware.
OpenOrganelle
The OpenOrganelle (Heinrich etal. 2021) from the Jane-
lia Research Campus hosts all of the datasets listed under
https:// openo rgane lle. janel ia. org/ datas ets on AWS’s Open
Data Registry. The data is visualizable in the browser using
neuroglancer or via Fiji N5-plugins. The data is currently
stored in a precursor to OME-Zarr known as the N5 for-
mat with “NGFF-compatible” metadata with conversion to
OME-Zarr on the future roadmap.
Fig. 15 Multiscale OME-Zarr representation of a whole brain dataset
(https:// downl oad. brain image libra ry. org/ 2b/ da/ 2bdaf 9e66a 246844/
mouse ID_ 405429- 182725/) (~ 6TB compressed) archived at BIL and
visualized over the internet in napari via the napari-ome-zarr plugin.
Data is stored at BIL in an alternative format and is dynamically con-
verted to OME-Zarr chunk-by-chunk and delivered to clients upon
request

Histochemistry and Cell Biology
1 3
Discussion
Over the last twenty years, the number of bioimaging file
formats has been a constant source of confusion and frustra-
tion. While that has often been a struggle that each user man-
ages in isolation, increasingly data sizes from more sophis-
ticated hardware and more advanced modalities are leaving
users with significant infrastructure burdens for efficiently
converting and sharing their imaging data. Data acquisi-
tion systems often design formats specifically for writing
data quickly to timely capture the scale and the breadth of
modern experiments. The tension between the requirements
of quickly writing and quickly reading bioimaging datasets
force both data providers and consumers to be aware of the
costs of converting, relinking, downsampling, or otherwise
modifying datasets for reuse. A one-time conversion of such
“write-optimized” data can lower the overhead of repeated
analysis and visualization of the data, but requires a widely
adopted target format. With proper support, a small suite of
storage file formats like HDF5, TIFF and Zarr can cover the
essential use cases for optimally achieving the community’s
scientific goals as has been achieved by other projects [e.g.,
PDB (Berman etal. 2012) and NetCDF (Unidata Ltd 1973)].
The strategy outlined in this paper is to encourage com-
munity cooperation towards a common representation.
Increased focus from the community of developers acceler-
ates the features delivered to the user community. Increases
in the expressive power of the format through specifications,
the number and ability of available tools, and data publicly
available for end users all motivate further developments in
each of the other areas. In turn, this progress drives the abil-
ity of the bioimaging community to better enact the FAIR
principles. The growth of OME-Zarr tools, resources, and
specifications, however, should not be taken as a reason
to wait on adoption. The opposite is true. The hope is that
more users and more developers will drive further growth,
further unifying the bioimaging ecosystem. Users should
identify whether or not the advancements detailed here will
simplify and accelerate their scientific practice, and if so, are
encouraged to start using OME-Zarr today. The community
is growing and membership is open, free and encouraged.
User feedback is critical to help make the most FAIR repre-
sentation of bioimaging data possible.
Author contributions Conceptualization: JM. Software: DB-L, SB, JB,
JB, EMB, J-MB, GdM, DG, SSG, IG, YOH, MH, DH, NH, MSK, GK,
AKY, KK, ML, TL, PL, DL, ML, JL, JM-S, TM, LM, MM, KM, JM,
WM, WO, BÖ, GP, CP, LP, TP, SP, NR, StS, SaS, NS, HS, ACS, DRS,
JS, CT, DT, IV, AMW, EW, KAY. Resources: SB, MH, KK, SO, MP.
Data curation: AKY, FW. Visualization: EED, SSG, GK, AlTdlV, JL,
MM, JM, WM, MP, CT, DT, AMW. Writing—original draft prepara-
tion: DB-l, SB, JB, EMB, J-MB, GdM, EED, SSG, IG, YOH, MH, DH,
MSK, GK, AKY, TL, PL, JL, JM-S, TM, MM, JM, BÖ, CP, LP, TP,
SP, NR, CT, DT, PW, AMW, FW, KAY. Writing—review and editing:
J-MB, XCM, BAC, YOH, NH, MSK, TM, JM, WM, NN, SP, NR, StS,
JS, JRS, EW. Supervision: OB, BAC, NG, MH, NH, SO, LAR, StS,
OS, JRS, FJT.
Funding J.M. was supported by Chan Zuckerberg Initiative DAF
for work on OME-NGFF by grant numbers 2019-207272 and 2022-
310144 and on Zarr by grant numbers 2019-207338 and 2021-237467.
S.S.G. and Y.O.H were supported by US National Institutes of Health
BRAIN Initiative award R24MH117295. The development of the Bio-
Image Archive has been supported by European Molecular Biology
Laboratory member states, Wellcome Trust grant 212962/Z/18/Z and
UKRI-BBSRC grant BB/R015384/1. The EMBL-EBI IT infrastructure
supporting the IDR and the BioImage Archive is funded by the UK
Research and Innovation Strategic Priorities Fund. M.K. was supported
by NHGRI 5T32HG002295. J.L was supported by grant 2022-252401
of the Chan Zuckerberg Initiative DAF, an advised fund of Silicon
Valley Community Foundation. T.M. was supported by the National
Science Foundation Graduate Research Fellowship under Grant No.
(DGE1745303). M.M. was supported by the US BRAIN Initiative
National Institutes of Health under award number 1RF1MH126732-
01. B.Ö. was supported by the EOSC Future project grant agreement
number: 101017536. S.O. was supported by JST NBDC Grant Number
JPMJND2201 and JST CREST Grant Number JPMJCR1926. N.A.H.,
N.J.S., S.B.S. and H.S. were funded via NCATS intramural research
fund. G.P. is supported by the Helmholtz Association under the joint
research school Munich School for Data Science and by the Joachim
Herz Foundation. L.M. is supported by the EMBL International PhD
Programme. M.L., J.Br., A.C.S. and L.A.R. were supported by the
Chan Zuckerberg Biohub San Francisco. N.N. was supported by Vin-
nova, grant number 2020-04702. T.P. was supported by grant number
2021-237557 from the Chan Zuckerberg Initiative DAF, an advised
fund of Silicon Valley Community Foundation. C.T. was funded by
grant number 2020-225265 from the Chan Zuckerberg Initiative DAF,
an advised fund of Silicon Valley Community Foundation. A.M.W was
supported by the US BRAIN Initiative National Institutes of Health
under award R24MH114793 and the Chan Zuckerberg Initiative for the
Brain Image Library Data Viewer Plugin Enhancement award 2022-
309651K.A.Y. was supported by the Open Research Data Program of
the ETH Board. Wellcome (Senior Clinical Research Fellowship, Well-
come Science Strategic Award) Work on OME-NGFF and IDR was
supported by the Wellcome Trust (ref. 212962/Z/18/Z), BBSRC (ref.
BB/R015384/1) and the National Institutes of Health Common Fund
4D Nucleome Program grant UM1HG011593. E.W. was supported by
Calico Life Sciences LLC; B.A.C was funded by NIH P41 GM135019,
and grant number 2020-225720 from the Chan Zuckerberg Initiative
DAF, an advised fund of Silicon Valley Community Foundation. O.W.
was supported by the SciLifeLab & Wallenberg Data Driven Life Sci-
ence Program (grant: KAW 2020.0239). N.G. was supported by NIH
OT2OD033758 and NIH R33CA263666.
Data availability All data shown in the figures is available publicly
under permissive licenses.
Code availability All code described is available publicly under free
and open-source licenses.
Declarations
Conflict of interest S.B., E.D., M.L., D.R.S. and J.R.S. are affiliated
with Glencoe Software, a commercial company that builds, deliv-
ers, supports and integrates image data management systems across
academic, biotech and pharmaceutical industries; J.M. and W.M. also
hold equity in Glencoe Software. M.M. is affiliated with Kitware, Inc.,
a commercial company built around open-source platforms that pro-
vides advanced technical computing, state-of-the-art AI, and tailored

Histochemistry and Cell Biology
1 3
software solutions to academic, government, and industrial customers.
A.V., J.S. and N.R. are affiliated with Scalable Minds, a commercial
company that builds, delivers, supports and integrates image analysis
solutions. F.J.T. consults for Immunai Inc., Singularity Bio B.V., Cy-
toReason Ltd, Cellarity, and Omniscope Ltd, and has ownership inter-
est in Dermagnostix GmbH and Cellarity. N.A.H. and N.J.S. are con-
tractors who work for Axle Research and Technology. S.B.S. and H.S.
are affiliated with Axle Research and Technology and are contracted to
the National Center for Advancing Translational Science, NIH. The re-
maining authors declare that they have no known competing financial
interests or personal relationships that could have appeared to influ-
ence the work reported.
Open Access This article is licensed under a Creative Commons Attri-
bution 4.0 International License, which permits use, sharing, adapta-
tion, distribution and reproduction in any medium or format, as long
as you give appropriate credit to the original author(s) and the source,
provide a link to the Creative Commons licence, and indicate if changes
were made. The images or other third party material in this article are
included in the article's Creative Commons licence, unless indicated
otherwise in a credit line to the material. If material is not included in
the article's Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a
copy of this licence, visit http:// creat iveco mmons. org/ licen ses/ by/4. 0/.
References
Ali HR, Jackson HW, Zanotelli VRT etal (2020) Imaging mass
cytometry and multiplatform genomics define the phenog-
enomic landscape of breast cancer. Nat Cancer 1:163–175.
https:// doi. org/ 10. 1038/ s43018- 020- 0026-6
Allan C, Burel J-M, Moore J etal (2012) OMERO: flexible, model-
driven data management for experimental biology. Nat Methods
9:245–253. https:// doi. org/ 10. 1038/ nmeth. 1896
Alted F (2010) Why modern CPUs are starving and what can be
done about it. Comput Sci Eng 12:68–71. https:// doi. org/ 10.
1109/ MCSE. 2010. 51
Bahry E, Breimann L, Zouinkhi M etal (2022) RS-FISH: precise,
interactive, fast, and scalable FISH spot detection. Nat Methods
19:1563–1567. https:// doi. org/ 10. 1038/ s41592- 022- 01669-y
Berman HM, Kleywegt GJ, Nakamura H, Markley JL (2012) The Pro-
tein Data Bank at 40: reflecting on the past to prepare for the
future. Structure 20:391–396. https:// doi. org/ 10. 1016/j. str. 2012.
01. 010
Besson S etal (2019) Bringing open data to whole slide imaging. In:
Reyes-Aldasoro C, Janowczyk A, Veta M, Bankhead P, Sirinuku-
nwattana K (eds) Digital pathology. ECDP 2019. Lecture notes in
computer science, vol 11435. Springer, Cham. https:// doi. org/ 10.
1007/ 978-3- 030- 23937-4_1
BICCN Data Ecosystem Collaboration, Hawrylycz MJ, Martone ME
etal (2022) The BRAIN initiative cell census network data eco-
system: a user’s guide. bioRxiv 2022.10.26.513573
Boergens KM, Berning M, Bocklisch T etal (2017) webKnossos: effi-
cient online 3D data annotation for connectomics. Nat Methods
14:691–694. https:// doi. org/ 10. 1038/ nmeth. 4331
Bogovic JA, Hanslovsky P, Wong A, Saalfeld S (2016) Robust reg-
istration of calcium images by learned contrast synthesis. In:
2016 IEEE 13th international symposium on biomedical imag-
ing (ISBI), pp 1123–1126. https:// doi. org/ 10. 1109/ ISBI. 2016.
74934 63
Bourget M-H, Kamentsky L, Ghosh SS etal (2022) Microscopy-BIDS:
an extension to the brain imaging data structure for microscopy
data. Front Neurosci 16:871228. https:// doi. org/ 10. 3389/ fnins.
2022. 871228
Bray M-A, Singh S, Han H etal (2016) Cell painting, a high-content
image-based assay for morphological profiling using multiplexed
fluorescent dyes. Nat Protoc 11:1757–1774. https:// doi. org/ 10.
1038/ nprot. 2016. 105
Casas Moreno X, Al-Kadhimi S, Alvelid J etal (2021) ImSwitch:
generalizing microscope control in Python. J Open Source Softw
6(64):3394. https:// doi. org/ 10. 21105/ joss. 03394
Casas Moreno X, Silva MM, Roos J etal (2023) An open-source
microscopy framework for simultaneous control of image acquisi-
tion, reconstruction, and analysis. HardwareX 13:e00400. https://
doi. org/ 10. 1016/j. ohx. 2023. e00400
Galaxy Community (2022) The Galaxy platform for accessible, repro-
ducible and collaborative biomedical analyses: 2022 update.
Nucleic Acids Res 50:W345–W351. https:// doi. org/ 10. 1093/ nar/
gkac2 47
de Boer P, Pirozzi NM, Wolters AHG etal (2020) Large-scale electron
microscopy database for human type 1 diabetes. Nat Commun
11:2475. https:// doi. org/ 10. 1038/ s41467- 020- 16287-5
de Boer IH, Alpers CE, Azeloglu EU etal (2021) Rationale and design
of the kidney precision medicine project. Kidney Int 99:498–510.
https:// doi. org/ 10. 1016/j. kint. 2020. 08. 039
Durbin C, Quinn P, Shum D (2020) Task 51—cloud-optimized format
study. https:// ntrs. nasa. gov/ citat ions/ 20200 001178
Granger BE, Pérez F (2021) Jupyter: thinking and storytelling with
code and data. Comput Sci Eng 23:7–14. https:// doi. org/ 10. 1109/
MCSE. 2021. 30592 63
Halchenko Y, Meyer K, Poldrack B etal (2021) DataLad: distributed
system for joint management of code, data, and their relationship.
J Open Source Softw 6:3262. https:// doi. org/ 10. 21105/ joss. 03262
Hartley M, Kleywegt G, Patwardhan A etal (2021) The BioIm-
age archive—home of life-sciences microscopy data. bioRxiv
2021.12.17.473169
Heinrich L, Bennett D, Ackerman D etal (2021) Whole-cell organelle
segmentation in volume electron microscopy. Nature 599:141–
146. https:// doi. org/ 10. 1038/ s41586- 021- 03977-3
Hörl D, Rojas Rusak F, Preusser F etal (2019) BigStitcher: recon-
structing high-resolution image datasets of cleared and expanded
samples. Nat Methods 16:870–874. https:// doi. org/ 10. 1038/
s41592- 019- 0501-0
HuBMAP Consortium (2019) The human body at cellular resolution:
the NIH Human Biomolecular Atlas Program. Nature 574:187–
192. https:// doi. org/ 10. 1038/ s41586- 019- 1629-x
Hunter (2007) Matplotlib: A 2D graphics environment. 9:90–95.
https:// doi. org/ 10. 1109/ MCSE. 2007. 55
Igarashi Y, Nakatsu N, Yamashita T etal (2015) Open TG-GATEs:
a large-scale toxicogenomics database. Nucleic Acids Res
43:D921–D927. https:// doi. org/ 10. 1093/ nar/ gku955
Kamentsky L, Jones TR, Fraser A etal (2011) Improved structure,
function and compatibility for Cell Profiler: modular high-
throughput image analysis software. Bioinformatics 27:1179–
1180. https:// doi. org/ 10. 1093/ bioin forma tics/ btr095
Keller MS, Gold I, McCallum C etal (2021) Vitessce: a framework
for integrative visualization of multi-modal and spatially-resolved
single-cell data. https:// doi. org/ 10. 31219/ osf. io/ y8thv
Könnecke M, Akeroyd FA, Bernstein HJ etal (2015) The NeXus data
format. J Appl Crystallogr 48:301–305. https:// doi. org/ 10. 1107/
S1600 57671 40275 75
Lange M, Granados A, VijayKumar S etal (2023) Zebrahub—multi-
modal Zebrafish developmental atlas reveals the state transition
dynamics of late vertebrate pluripotent axial progenitors. bioRxiv
2023.03.06.531398

Histochemistry and Cell Biology
1 3
Lim I, Yu Lin E, Garcia J etal (2023) Shortwave infrared fluorofluoro-
phores for multicolor invivo imaging. Angew Chem Int Ed Engl
62:e202215200. https:// doi. org/ 10. 1002/ anie. 20221 5200
Linkert M, Rueden CT, Allan C etal (2010) Metadata matters: access
to image data in the real world. J Cell Biol 189:777–782. https://
doi. org/ 10. 1083/ jcb. 20100 4104
Long B, Miller J, The SpaceTx Consortium (2023) SpaceTx: a road-
map for benchmarking spatial transcriptomics exploration of the
brain. arXiv: 2301. 08436 v1
Unidata Ltd (1973) Unidata. Comput Aided Des 5:48. https:// doi. org/
10. 1016/ 0010- 4485(73) 90157-7
Major B, McCormick M (2022) KitwareMedical/tensorboard-plugin-
3d: v1.0.3. https:// doi. org/ 10. 5281/ zenodo. 65222 67
Manz T, Gold I, Patterson NH etal (2022) Viv: multiscale visualization
of high-resolution multiplexed bioimaging data on the web. Nat
Methods. https:// doi. org/ 10. 1038/ s41592- 022- 01482-7
Marconato L, Palla G, Yamauchi KA, Virshup I, Heidari E, Treis T,
Toth M, Shrestha RB, Vöhringer H, Huber W, Gerstung M, Moore
J, Theis FJ, Stegle O (2023) SpatialData: an open and universal
data framework for spatial omics. bioRxiv. https:// doi. org/ 10.
1101/ 2023. 05. 05. 539647
McCormick M (2022) itk-wasm: high-performance spatial analysis in
a web browser, Node.js, and reproducible execution across pro-
gramming languages and hardware architectures. https:// doi. org/
10. 5281/ zenodo. 74749 40
McCormick M, Liu X, Jomier J etal (2014) ITK: enabling reproducible
research and open science. Front Neuroinform 8:13. https:// doi.
org/ 10. 3389/ fninf. 2014. 00013
McCormick M, Major B, Abdala L etal (2022) InsightSoftwareCon-
sortium/itkwidgets: itkwidgets 1.0 Alpha 21. https:// doi. org/ 10.
5281/ zenodo. 73531 49
McDole K, Guignard L, Amat F etal (2018) In toto imaging and recon-
struction of post-implantation mouse development at the single-
cell level. Cell 175:859-876.e33. https:// doi. org/ 10. 1016/j. cell.
2018. 09. 031
Miles A, jakirkham, Bussonnier M etal (2023) zarr-developers/zarr-
python: v2.15.0 Zenodo. https:// doi. org/ 10. 5281/ zenodo. 80391 03
Moore J, Allan C, Besson S etal (2021) OME-NGFF: a next-gen-
eration file format for expanding bioimaging data-access strat-
egies. Nat Methods 18:1496–1498. https:// doi. org/ 10. 1038/
s41592- 021- 01326-w
Nelson G, Boehm U, Bagley S etal (2021) QUAREP-LiMi: a commu-
nity-driven initiative to establish guidelines for quality assessment
and reproducibility for instruments and images in light micros-
copy. arXiv. 2101. 09153
Ouyang W, Mueller F, Hjelmare M etal (2019) ImJoy: an open-source
computational platform for the deep learning era. Nat Methods
16:1199–1200. https:// doi. org/ 10. 1038/ s41592- 019- 0627-0
Ouyang W, Beuttenmueller F, Gómez-de-Mariscal E, etal (2022) Bio-
Image model zoo: a community-driven resource for accessible
deep learning in bioimage analysis. bioRxiv 2022.06.07.495102
Pape C, Meechan K, Moreva E etal (2022) MoBIE: a Fiji plugin for
sharing and exploration of multi-modal cloud-hosted big image
data. bioRxiv 2022.05.27.493763
Payne AC, Chiang ZD, Reginato PL etal (2021) Insitu genome sequenc-
ing resolves DNA sequence and structure in intact biological sam-
ples. Science. https:// doi. org/ 10. 1126/ scien ce. aay34 46
Perez F, Granger BE (2007) IPython: a system for interactive scientific
computing. Comput Sci Eng 9:21–29. https:// doi. org/ 10. 1109/
mcse. 2007. 53
Pietzsch T, Saalfeld S, Preibisch S, Tomancak P (2015) BigData-
Viewer: visualization and processing for large image data sets.
Nat Methods 12:481–483. https:// doi. org/ 10. 1038/ nmeth. 3392
Preibisch S, Karaiskos N, Rajewsky N (2022) Image-based repre-
sentation of massive spatial transcriptomics datasets. bioRxiv
2021.12.07.471629
Ramachandran R, Bugbee K, Murphy K (2021) From open data to open
science. Earth Space Sci. https:// doi. org/ 10. 1029/ 2020e a0015 62
Rueden CT, Ackerman J, Arena ET etal (2019) Scientific Community
Image Forum: a discussion forum for scientific image software.
PLoS Biol 17:e3000340. https:// doi. org/ 10. 1371/ journ al. pbio.
30003 40
Rzepka N, Bogovic JA, Moore JA (2023) Toward scalable reuse of
vEM data: OME-Zarr to the rescue. Methods in cell biology. Aca-
demic Press.https:// doi. org/ 10. 1016/ bs. mcb. 2023. 01. 016
Sarkans U, Chiu W, Collinson L etal (2021) REMBI: recommended
metadata for biological images-enabling reuse of microscopy data
in biology. Nat Methods 18:1418–1422. https:// doi. org/ 10. 1038/
s41592- 021- 01166-8
Schapiro D, Yapp C, Sokolov A etal (2022) MITI minimum informa-
tion guidelines for highly multiplexed tissue images. Nat Methods
19:262–267. https:// doi. org/ 10. 1038/ s41592- 022- 01415-4
Schindelin J, Arganda-Carreras I, Frise E etal (2012) Fiji: an open-
source platform for biological-image analysis. Nat Methods
9:676–682. https:// doi. org/ https:// doi. org/ 10. 1038/ nmeth. 2019
Sofroniew N, Lambert T, Evans K etal (2022) napari: a multi-dimen-
sional image viewer for Python. Zenodo. https:// doi. org/ 10. 5281/
ZENODO. 35556 20
Tohsato Y, Ho K, Kyoda K, Onami S (2016) SSBD: a database of quan-
titative data of spatiotemporal dynamics of biological phenomena.
Bioinformatics. https:// doi. org/ 10. 1093/ bioin forma tics/ btw417
Valuchova S, Mikulkova P, Pecinkova J etal (2020) Imaging plant
germline differentiation within Arabidopsis flowers by light sheet
microscopy. Elife. https:// doi. org/ 10. 7554/ eLife. 52546
Vergara HM, Pape C, Meechan KI etal (2020) Whole-body integra-
tion of gene expression and single-cell morphology. Cold Spring
Harbor Laboratory. https:// doi. org/ 10. 1101/ 2020. 02. 26. 961037
Virshup I, Rybakov S, Theis FJ etal (2021) anndata: Annotated data.
bioRxiv 2021.12.16.473007
Way GP, Natoli T, Adeboye A etal (2022) Morphology and gene
expression profiling provide complementary information for map-
ping cell state. Cell Syst 13:911-923.e9. https:// doi. org/ 10. 1016/j.
cels. 2022. 10. 001
Weisbart E, Cimini BA (2022) Distributed-Something: scripts to lev-
erage AWS storage and computing for distributed workflows at
scale. arXiv. 2210. 01073
Wilkinson MD, Dumontier M, Aalbersberg IJJ etal (2016) The FAIR
guiding principles for scientific data management and steward-
ship. Sci Data 3:160018. https:// doi. org/ 10. 1038/ sdata. 2016. 18
Williams E, Moore J, Li SW etal (2017) The image data resource: a
bioimage data integration and publication platform. Nat Methods
14:775–781. https:// doi. org/ 10. 1038/ nmeth. 4326
Publisher's Note Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations.
Histochemistry and Cell Biology
1 3
Authors and Aliations
JoshMoore
1
· DanielaBasurto‑Lozada
2
· SébastienBesson
3
· JohnBogovic
4
· JordãoBragantini
5
·
EvaM.Brown
6
· Jean‑MarieBurel
3
· XavierCasasMoreno
7
· GustavodeMedeiros
8
· ErinE.Diel
9
·
DavidGault
3
· SatrajitS.Ghosh
10
· IlanGold
11
· YaroslavO.Halchenko
12
· MatthewHartley
13
·
DaveHorsfall
2
· MarkS.Keller
11
· MarkKittisopikul
4
· GaborKovacs
14
· AybükeKüpcüYoldaş
13
·
KojiKyoda
15
· AlbaneleTournoulxdelaVillegeorges
16
· TongLi
17
· PriscaLiberali
8
· DominikLindner
3
·
MelissaLinkert
9
· JoelLüthi
8
· JeremyMaitin‑Shepard
18
· TrevorManz
11
· LucaMarconato
19
·
MatthewMcCormick
20
· MerlinLange
5
· KhaledMohamed
3
· WilliamMoore
3
· NilsNorlin
21
·
WeiOuyang
7
· BugraÖzdemir
22
· GiovanniPalla
23
· ConstantinPape
24
· LucasPelkmans
25
·
TobiasPietzsch
4
· StephanPreibisch
4
· MartinPrete
17
· NormanRzepka
16
· SameeulSamee
26
·
NicholasSchaub
27
· HythemSidky
26
· AhmetCanSolak
5
· DavidR.Stirling
9
· JonathanStriebel
16
·
ChristianTischer
28
· DanielToloudis
6
· IsaacVirshup
23
· PetrWalczysko
3
· AlanM.Watson
29
·
ErinWeisbart
30
· FrancesWong
3
· KevinA.Yamauchi
31
· OmerBayraktar
17
· BethA.Cimini
30
·
NilsGehlenborg
11
· MuzlifahHania
17
· NathanHotaling
27
· ShuichiOnami
15
· LoicA.Royer
5
·
StephanSaalfeld
4
· OliverStegle
19
· FabianJ.Theis
23
· JasonR.Swedlow
3
* Josh Moore
josh@openmicroscopy.org
1
German BioImaging-Gesellschaft für Mikroskopie und
Bildanalyse e.V., Constance, Germany
2
Biosciences Institute, Newcastle University,
NewcastleuponTyne, UK
3
Divisions ofMolecular Cell andDevelopmental Biology,
andComputational Biology, University ofDundee, Dundee,
Scotland,UK
4
Janelia Research Campus, Howard Hughes Medical Institute,
Ashburn, VA, USA
5
Chan Zuckerberg Biohub, SanFrancisco, CA, USA
6
Allen Institute forCell Science, Seattle, WA, USA
7
Science forLife Laboratory, KTH Royal Institute
ofTechnology, Stockholm, Sweden
8
Friedrich Miescher Institute forBiomedical Imaging, Basel,
Switzerland
9
Glencoe Software Inc., Seattle, WA, USA
10
Massachusetts Institute ofTechnology, Cambridge, MA,
USA
11
Harvard Medical School, Boston, MA, USA
12
Dartmouth College, Hanover, NH, USA
13
European Molecular Biology Laboratory, European
Bioinformatics Institute, EMBL-EBI, Cambridge, UK
14
Allen Institute forNeural Dynamics, Seattle, WA, USA
15
RIKEN Center forBiosystems Dynamics Research, Kobe,
Japan
16
scalable minds GmbH, Potsdam, Germany
17
Wellcome Sanger Institute, Hinxton, UK
18
Google Research, MountainView, CA, USA
19
Genome Biology Unit, European Molecular Biology
Laboratory (EMBL), Heidelberg, Germany
20
Kitware, Inc, Carrboro, NC, USA
21
Department ofExperimental Medical Science & Lund
Bioimaging Centre, Lund University, Lund, Sweden
22
Euro-BioImaging Bio-Hub, EMBL, Heidelberg, Germany
23
Institute ofComputational Biology, Helmholtz Zentrum
München, Neuherberg, Germany
24
Georg-August-Universität Göttingen, Göttingen, Germany
25
University ofZürich, Zürich, Switzerland
26
Axle Research andTechnology, Rockville, USA
27
Information Technology Branch, National Center
forAdvancing Translational Science, National Institutes
ofHealth, Bethesda, USA
28
EMBL, Heidelberg, Germany
29
University ofPittsburgh, Pittsburgh, PA, USA
30
Imaging Platform, Broad Institute ofMIT andHarvard,
Cambridge, MA, USA
31
Department ofBiosystems Science andEngineering, ETH
Zürich, Zürich, Switzerland
